Survivin Inhibitors as a Target Therapy for Leukemia
Download
Abstract
Leukemia is a type of cancer characterized by the accumulation of leukemic cells in the patient’s peripheral blood. Traditional therapies often fail to completely eliminate these cancer cells, especially in refractory leukemia. One major reason for this failure is apoptosis dysfunction, with anti-apoptotic proteins playing crucial roles in cancer cell survival. Among these proteins, the inhibitor of apoptosis proteins (IAPs), particularly survivin, are highly overexpressed in most cancers, including leukemia, and contribute to chemotherapy resistance. Survivin, an inhibitor of apoptosis, is typically expressed during embryonic development and in tumor cells but not in normal adult tissues. It suppresses apoptosis, thereby promoting disease progression and resistance to chemotherapy. In leukemia, survivin expression is associated with poor prognostic outcomes in both acute myeloid leukemia (AML) and chronic lymphocytic leukemia (CLL). The dysregulation of apoptosis in leukemic cells is often linked to increased survivin expression, making it a promising therapeutic target. This review explores the diverse roles of survivin in mediating apoptosis, cell division, and chemoresistance in leukemia. It also discusses several survivin-targeting strategies, including ribozymes, immunotherapy, and gene therapy. Preclinical and clinical studies involving survivin inhibitors are currently underway and show promise in increasing the sensitivity of leukemic cells to standard chemotherapy treatments. These therapies, by disrupting survivin’s protective mechanisms, may lead to better treatment outcomes for leukemia patients. Understanding the complex relationship between survivin and apoptotic pathways will provide valuable insights for developing innovative therapeutic strategies against these challenging hematologic malignancies.
Introduction
Leukemia is a type of cancer that affects white blood cells, accounting for about 8% of cancers across various age groups worldwide [1]. It is characterized by an accumulation of leukemic cells in the peripheral blood due to disturbances in cell proliferation and differentiation [2]. While conventional therapies effectively reduce the high proliferative population of tumor cells, refractory leukemia remains a significant therapeutic challenge. Multiple factors contribute to the ineffectiveness of many treatments including genetic abnormalities [3], drug resistance [4], age and sex [5]. Leukemias may present at all ages, from the newborn to the very old, with different forms [6]. According to evidence, the prevalence of ALL is most common in early childhood, whereas AML is increasingly common in older adults. Eighty percent of hematological malignancies in children are ALL, while the others (15%–20%) include AML and hematology-related diseases. Additionally, the response rate to treatment is significantly higher in the younger age group than in older patients [7].
Comprehensive studies on the incidence of AML and ALL in children and older patients show that the incidence of AML is 1.5 per 100,000 individuals per year in children aged 1-9, while its prevalence in people over 65 years old is 6.5 per 100,000 per year [8].
Germ-line mutations in several genes such as TP53, RUNX1, and GATA2, as well as some genetic aberrations, have different prognostic impacts on adult and pediatric patients with AML [9].
Patients older than 56 years have an increased incidence of unfavorable cytogenetics, which is associated with a poor prognosis [10, 11]. Comprehensive studies on the incidence of AML and ALL in children and older patients show that the incidence of AML is 1.5 per 100,000 individuals per year in children aged 1-9, while its prevalence in people over 65 years old is 6.5 per 100,000 per year [8].
Germ-line mutations in several genes such as TP53, RUNX1, and GATA2, as well as some genetic aberrations, have different prognostic impacts on adult and pediatric patients with AML [9].
Patients older than 56 years have an increased incidence of unfavorable cytogenetics, which is associated with a poor prognosis [10, 11]. ALL, defined by an excess of lymphoblasts of either the B or T lineage, is more prevalent in males than in females and varies in incidence among ethnic groups, with Hispanics having the greatest prevalence [20, 21]. Several genetic variables, including mutations linked to Down’s syndrome, PAX5, and ETV6, as well as polymorphism variations in particular genes such as ARID5B, CEBPE, GATA3, and IKZF1, are associated with an elevated risk of ALL [12].
Studies indicate that CLL is the most common adult leukemia in the Western world, accounting for 25% of adult leukemias and non-Hodgkin’s lymphoma (NHL). The median age of diagnosis for chronic lymphocytic leukemia (CLL) in the United States is 71 years, with an adjusted incidence rate of 4.5 cases per 100,000 individuals [21].
Huang et al. predicted that the prevalence of CML in the United States will rise from around 70,000 in 2010 to a near plateau of 35 times the yearly incidence by 2050 [13]. This abnormal kinase signaling activates downstream targets, causing the reprogramming of cells and resulting in uncontrolled proliferation, myeloid hyperplasia, and the development of ‘indolent’ symptoms in the chronic phase (CP) of CML [14].
Given the widespread occurrence of leukemia across all age groups, ranging from newborns to elderly individuals, it is imperative to acquire knowledge about genetic and molecular components in order to discover novel treatments for all leukemia patients [3].
One of the key factors is the disruption of the apoptotic process [15]. The development of different hematological malignancies, such as leukemia, depends on an imbalance between cell proliferation and death [16]. Various anomalies in apoptosis regulation systems result in resistance to caspase-dependent cell death, often associated with increased expression of proteins that inhibit apoptosis [17]. Among the anti-apoptotic proteins, inhibitor of apoptosis proteins (IAPs) play a crucial role. They specifically block the activity of caspase 3 and 7 thereby preventing apoptosis.
A stude by Olga Grzybowska-Izydorczyk revealed that higher expression of IAPs including cIAP1 and cIAP2 is related to lower survival rates or poor responses to chemotherapy in chronic lymphocytic leukemia (CLL) and non-Hodgkin’s lymphoma [18, 19].
Survivin is a unique member of the IAP family that is selectively overexpressed in almost all human cancer cells, especially in leukemia [20]. Previous studies also indicate that survivin has low expression in most normal adult tissues [21]. Therefore, apoptosis inhibition might be a common feature of neoplasia, contributing to disease progression and resistance to therapy [22-24]. This review focuses on significant findings regarding survivin expression in various malignancies, including leukemia, and the potential therapeutic applications of survivin inhibition and its clinical implications.
3- Inhibitor of apoptosis proteins (IAP) in leukemia
The fundamental occurrence in apoptosis is the activation of caspases through both the death receptor (external) and mitochondrial (intrinsic) pathways. This triggers DNA fragmentation, the cleavage of many essential proteins, and ultimately the disintegration of the cell [25]. Apoptosis, the process of programmed cell death, is controlled by many regulatory systems operating at different levels. Regarding to leumeia, IAPs refer to inhibitor of apoptosis proteins specially caspases, they play a fundamental role in preventing apoptosis and contribute to the cell survival [26, 27]. Thay have at least one copy of Baculovirus IAP Repeat (BIR) domain in their N-terminal portion. The IAPs family containes 8 members including NAIP, cIAP1, cIAP2, XIAP, Survivin, Bruce/Apollon, ML-IAP/Livin and ILP-2. In the first IAPs were discovered in viruses and owing to their function in the viruses, received the name of IAPs [28]. Survivin as a most important member of IAPs has a critical role in the repression of apotosis. The importance of survivin among the IAPs is related to high expression in various cancers and interaction with fundamental apoptosis related proteins and consequently the preogression of cancer [29]. Research has proven that targeting survivin may increase the effectiveness of anticancer agents. Thus, survivin has become an important focus for developing novel treatments aimed at improving patient outcomes in cancer [29].The overexpression of IAPs has been observed in various cancer cell lines and primary tumor biopsy samples, including leukemic cell lines or blasts isolated from patients with acute leukemia. The potential role of these proteins is currently being evaluated for their involvement in defective apoptotic regulatory mechanisms, leukemic clone progression, and the development of chemoresistance [30-32]. The most potent and well-known members of this family in humans are XIAP, c-IAP1, c-IAP2, and survivin [28].
Most IAP operations rely on BIR domains. The BIR2 domain of IAPs has the ability to attach to and inhibit caspases-3 and -7, whereas BIR3 acts as an inhibitory segment for caspase-9. BIR1 does not possess the capacity to prevent caspase activation [28]. Both cIAP1 and cIAP2 have the ability to hinder the enzymatic function of caspase-3, -7, and -9; however, they are not as powerful as XIAP [28]. BIR domains are involved in the contact between cIAP1 and cIAP2 with TNF receptor-related factors (TRAF1 and TRAF2), as well as the interaction between XIAP and TAB1 protein. This interaction leads to the activation of the nuclear factor κB (NF-κB) pathway [33]. Many studies have demonstrated that reducing the activity of IAP can potentially overcome the resistance of leukemic cells to chemotherapy. In the case of HL-60 cells, when treated with XIAP antisense oligonucleotide along with different concentrations of cytosine arabinoside (Ara-C), the down-regulation of XIAP led to the activation of caspases and increased the susceptibility of leukemic cells to Ara-C-induced cell death [34]. Hundsdoerfer et al. have discovered that children with acute lymphoblastic leukemia (ALL) cells have a high expression of XIAP protein compared to bone marrow mononuclear cells. This high expression of XIAP is associated with a poor response to prednisone treatment in children with T-cell ALL. These findings suggest that XIAP might be an important target for therapy in pediatric ALL [35].
This proves that IAP inhibitors synergize with numerous cytotoxic medications routinely used in the treatment of pediatric ALL to induce apoptosis and prevent long-term clonogenic survival [35]. Research by Löder .S suggests that using IAP inhibitors in chemotherapy regimens might minimize the dosage of cytotoxic medicines needed for antileukemic action, potentially reducing the risk of chemotherapy-related harmful side effects [36]. Beyond pediatric ALL, the proposed molecular mechanism of synergy has larger implications for other malignancies and offers new avenues for the use of inhibitors in chemotherapy-based combinations.
IAP inhibitors have just reached phase 1 clinical trials, and combinations with standard chemotherapeutic drugs may quickly be put into clinical practice [37-40] . Thus, IAP inhibitors, either alone or in combination with chemotherapy, represent a potential new method for apoptosis-targeted therapeutics for not just pediatric ALL, but all leukemias.
4- Survivin
Survivin is the smallest member of the IAP (Inhibitor of Apoptosis Protein) family and is involved in controlling the cell cycle and preventing apoptosis. Its expression patterns vary; it is predominantly expressed during the embryonic phase and found in differentiated tissues and cancer cells but not in normal adult cells. In cancer tumors, survivin expression inhibits apoptosis, reduces cell death, resists chemotherapy, and promotes tumor invasion [41]. Survivin has a single baculovirus IAP repeat (BIR) domain and lacks a carboxyl-terminal RING finger. It is highly expressed in all common human malignancies, including lung, colon, and pancreatic cancers, and in high-grade non-Hodgkin lymphomas, but not in lower- grade lymphomas [42]. Survivin also plays a role in angiogenesis, which can contribute to the inhibition of apoptosis [43].
4-1. The apoptotic pathway of survivin
Survivin can activate cell death in several ways. External pathways involve ligand binding (e.g., FasL, TNF) to cell surface receptors, while internal pathways include direct mitochondrial signaling. The activation of the mitochondrial pathway occurs through the action of Bax/Bcl-2, resulting in the release of apoptotic components like cytochrome c into the cytoplasm [39].
This triggers the formation of a complex known as the cytochrome c/Apaf-1/caspase-9 apoptosome, which activates caspase-3, leading to the breakdown of skeletal proteins and DNA. Inhibitors of apoptosis, such as survivin, prevent apoptosis through both caspase- dependent and non-caspase-dependent pathways [44].
4-2. Survivin in Acute Myeloid Leukemia (AML)
AML is a heterogeneous and malignant disease characterized by a significant increase in the growth of immature leukocytes. Survivin, an anti-apoptotic protein, is detectable in leukemia [45].
The level of survivin in CD34+38− AML stem cells is higher than in larger blasts and in all CD34+ AML cells. Survivin expression correlates with several proteins involved in cell proliferation and survival, making it a predictive biomarker and a key target for cancer treatment in AML [41].
Targeting survivin with antisense oligonucleotides induces cellular proliferation and subsequent cell death in AML cells [46].
4-3. Survivin in B-Cell Precursors and ALL
Survivin is highly expressed in two-thirds of B-cell precursors, unlike normal blood cells. Deleting survivin via short-hairpin RNA or antisense oligonucleotides promotes death in leukemia cells [47]. Survivin-Ex3/WT expression levels are used to classify children with B-cell ALL into high-risk and low-risk groups [48]. In chronic lymphocytic leukemia (CLL), insufficient apoptosis results in the accumulation of CD5+ B cells in lymphatic organs, bone marrow, and peripheral circulation.
The expression and regulation of the apoptotic inhibitory protein family are critical in the treatment of leukemia. B cells in the microenvironment can use physiological inputs like the CD4 ligand to regulate the production of apoptotic inhibitors.
In vitro findings from peripheral blood and bone marrow mononuclear cells from various patients show that B cells stimulate CD4, and survivin is the only apoptosis-inducing protein produced by CD4. In vivo, survivin expression was assessed using immunohistochemistry in cervical reactive lymph nodes located in the proliferative germinal core. Patients with B-cell survival had only pseudofollicles found in their lymph nodes. Pseudofollicles containing survivin+ cells were active in cell proliferation. Unlike the survivin+ B cells detected in the germinal core, they were BCL2+. CD5+ survivin cells are found in clusters densely packed with T cells in B cells from bone marrow biopsy samples. These findings demonstrate that survivin governs B cell proliferation and that its expression can be modulated by environmental cues [49].
4-4. Survivin in T-Cell Leukemia and Other Leukemias
Survivin is consistently expressed in all ATL (Adult T-cell Leukemia) patients, making survivin mRNA measurement suitable for real-time PCR. Survivin levels serve as a biomarker for clinical stages or minimal residual disease (MRD) [50]. Survivin may be detected in all myeloid leukemia cell lines using Western blot analysis, but not in normal CD34+ cells and peripheral blood mononuclear cells. Cytokine stimulation increases survivin expression in leukemia cell lines and primary AML samples. In contrast, all-trans retinoic acid significantly lowers survivin protein levels, leading to myelomonocytic differentiation [46].
4-5. Regulation and Therapeutic Potential
The MEK (mitogen-activated protein kinase) and PI3K (phosphatidylinositol-3-kinase) pathways play distinct roles in regulating survivin expression. These findings indicate that survivin is cytokine-regulated in myeloid leukemia and is substantially expressed in various leukemias, suggesting that hematopoietic cytokines partially mediate their antitumor and mitogenic actions. Targeting survivin, either alone or in combination with chemotherapy, represents a promising approach for treating leukemia by promoting apoptosis and reducing chemoresistance [51].
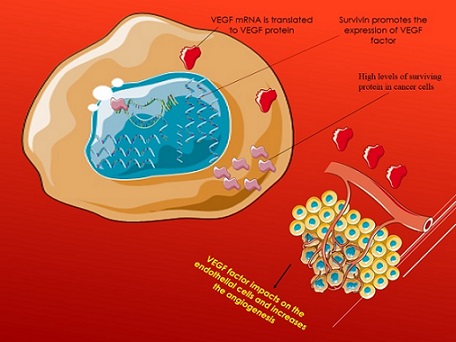
Binding of surviving/VEGF in angiogenesis
Survivin-expressing tumor cells promote the synthesis and secretion of VEGF via the B-catenin signal. Along with the VEGF secreted by endothelial cells, the released VEGF may operate on endothelial cells to promote angiogenesis. On the other hand, in tumors with low survivin cells, it stimulates VEGF synthesis and secretion, which may enhance vasoconstriction [52].
Survivin and VEGF protein expression were analyzed using reverse transcription (RT)-PCR and western blotting, respectively. In addition, survivin and VEG concentrations were measured using an enzyme-linked immunosorbent test (ELISA). Survivin and VEGF were overexpressed in ALL patients before treatment, whereas survivin levels were dramatically reduced after treatment. In addition, there was a positive association between survivin and VEGF levels in plasma [53] (Figure 1).
Figure 1. The Interaction between Surviving Protein and VEGF Factor in Cancer Cell Metastasis.
4-1. Molecular Organization and Structure of Survivin
Survivin is a key member of the IAP family. Unlike other IAPs, it features only one N-terminal BIR domain, a lengthy C-terminal alpha-helix coiled region, and a dimeric structure. The BIR domain is assumed to be crucial for antiapoptotic action, whereas the coilcoiled domain most likely interacts with tubulin structures [54, 55]. Survivin has three chemically different surfaces: acidic and basic areas, on the BIR domain and a hydrophobic helical surface on alpha 6 [55]. This organization corresponds to functionally significant protein-protein interaction surfaces. Survivin is encoded by the BIRC5 gene on chromosome 17q25 and produces transcripts with a variety of functional domains. The BIRC5 gene produces wild-type Survivin WT and five splice variants: Ex3, 2B Survivin, 3B, 2, and 3 [56, 57]. Survivin and its splice variants are also localized differently within cells. Survivin and survivin-2B are cytoplasmic proteins, while survivin-ΔEx-3 is mostly nuclear. Survivin isoforms and their various sites in the cell may suggest a regulatory balance between apoptosis and suppression of apoptosis [58, 59].
4-2. Survivin performance in cell division
Survivin is mostly increased during the G2/M phase of the cell cycle in a manner that depends on the cell cycle homology area within the promoter. This suggests that survivin may assist cancer cells in bypassing the G2/M checkpoint, hence promoting unlimited cell proliferation [60, 61]. The localization of survivin during mitosis is in accordance with the presence of chromosomal passenger proteins (CPP) such as Aurora B, Borealin, and inner centromere protein (INCENP) [62]. Survivin accumulates at centrosomes during early mitosis, specifically at mitotic histone marks. This accumulation is facilitated by the phosphorylation of Thr3 on histone 3 (H3) by Haspin-mediated process [57].
Survivin binds to phosphorylated H3 and then guides its CPP partners to inner centromeres, resulting in the formation of a chromosomal passenger complex (CPC). Aurora B kinase, the CPC’s enzymatic subunit, corrects syntelic and merotelic defects in kinetochore microtubule attachment, ensuring equitable sister chromatid distribution [59-61]. Survivin and its CPP partners disassociate from kinetochores during anaphase, but in telophase, they re-aggregate at the polar end of microtubules [63-65]. At this site, the CPC enzyme phosphorylates proteins that control the contractile actin-myosin ring, including MgcRacGAP and SHC SH2-domain binding protein 1 (SHCBP1) [65].
Furthermore in this process survivin interacts with tubulin, that localizes to microtubule organizing centers (MTOC) during mitosis. The localization of survivin to the spindle apparatus is functionally associated with its capacity to inhibit the apoptotic signal controlled by the TNF receptor family member, Fas, and the death cascade triggered by the release of cytochrome c from mitochondria produced by Bax [65]. Depletion of survivin causes defective cell division that involves activation of spindle checkpoints mediated by tumor suppressor protein p53 due to an arrest of DNA synthesis. Survivin deficient cells frequently fail to complete both chromosome segregation and cytokinesis during mitosis. In the absence of survivin, sister chromatids begin to separate normally during anaphase but frequently fail to follow the main mass of segregating chromosomes, resulting in aberrant chromatid separation [66, 67]. Without survivin, cytokinesis fails in late stages due to aberrant spindle midzone and midbody microtubule production. A faulty CPC has been shown to cause problems in chromosomal segregation and cytokinesis. These studies show that Survivin plays an important role in mitosis and cell division [65-67] (Table 1).
| Involved molecules | Role of molecules | References |
| Chromosomal passenger proteins (CPP) | localization of survivin during mitosis | 57 |
| Inner centromere protein (INCENP) | localization of survivin during mitosis | 57 |
| histone 3 (H3) | Accumulation of surviving in centrosomes during early mitosis | 59-61 |
| Aurora B kinase | It is CPC's enzymatic subunit, ensuers equitable sister chromatid distribution | 59-61 |
| Chromosomal passenger complex (CPC) enzyme | phosphorylates proteins that control the contractile actin-myosin ring, including MgcRacGAP and SHC SH2-domain binding protein 1 (SHCBP1). | 60 |
| Tubulin | The interaction of surviving with tubulin has direct relation with surviving capacity in the inhibition of apoptosis. | 60 |
4-3. Survivin performance in apoptosis
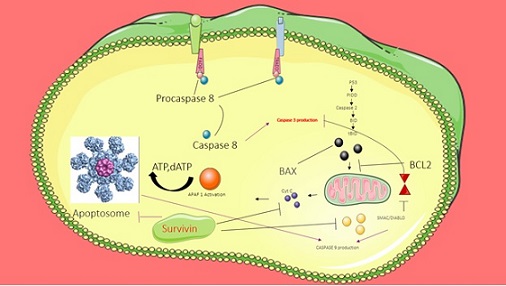
Survivin prevents apoptosis through two distinct pathways: caspase-dependent (external) and mitochondrial (interior). Studies have revealed suppression of these caspases via direct physical interaction between survivin and caspases 3 and 9, if direct binding of survivin is suspected, or indirect inhibition via hepatitis B-interacting protein (HBXIP) for caspase 9 [68, 69].
In Following the activation of initiator caspase-8, death receptors (CD-95/Fas and TNF receptors) are activated via external signals, hence initiating the extrinsic apoptotic cascade. On the other hand. Intracellular signals acting through mitochondria trigger the intrinsic apoptosis process. In response to signals, mitochondria release cytochrome-c (cyt-c) and Smac/DIABLO, forming apoptosomes that activate the initiator caspase-9 [69, 70]. Survivin has been demonstrated to be abundant in the inter-mitochondrial membrane space. The mechanism underlying survivin localization in mitochondria is yet unknown. However, the molecular chaperone heat shock protein 90 (Hsp90) is thought to help import several client proteins to mitochondria and has been linked to survivin. [71] . Smac/DIABLO operates as a pro-apoptotic protein because it participates in the development of an apoptosome and activates caspase-9. Smac/DIABLO has been reported to antagonize XIAP functioning [72]. Thus, the presence of survivin may indirectly allow XIAP to function, ultimately inhibiting apoptosis (Figure 2).
Figure 2. The Role of Surviving Protein in the Regulation of Apoptosis.
4-4 Survivin performance in cancer drug resistance
Chemotherapy is crucial in the treatment of cancer patients. However, therapeutic resistance is one factor that lowers therapy response. Drug resistance follows the typical method of drug export from cells, which involves the overexpression of P-glycoprotein (P-gp) and ABC transporters [73]. Survivin is implicated in the resistance to chemotherapy [74, 75]. Survivin expression has been revealed to be very high after treatment with chemotherapy drugs in cancer cells [76, 77]. Survivin transcription has been also reported to be associated with the P-gp / MDR1 overexpression [78]. However, in the HL60 cell line, the P-gp does not appear to play a significant role as a specific inhibitor of apoptosis proteins (IAPs). The expression of p53 or BcL-2 has been found to be similar in HL60 and HL60R cells. However, in HL60R cells, mRNA levels of IAPs such as survivin, c-IAP2, and NAIP have been shown to increase. In addition, doxorubicin therapy has been shown to drastically reduce X-linked IAP and survivin mRNA levels in HL60 cells. Cisplatin has also been shown to have lower impact on survivin and NAIP mRNA levels in the same cells. However, in HL60R cells, it has been shown that these mRNA levels are less influenced by the therapies. As a result, these data show that IAPs may be involved in tumor resistance to chemotherapeutic drugs [79]. Moreover, survivin mRNA expression in human leukemia K-562/ADR cell line with acquired resistance to adriamycin has been illustrated to be 1.7 times higher than in K-562 cells [80]. These studies suggest that survivin can be a potential target in the treatment of hematopoietic cancers (HCs) [20]. Patients with undetectable survivin levels demonstrated full remissions, whereas patients resistant to imatinib presented greater survivin levels [81]. Furthermore, survivin levels were shown to be considerably lower in imatinib-sensitive CML cells. Imatinib inhibits the tyrosine kinase activity of the oncogene BCR-ABL in CML cells, lowering the production of cellular proteins and tumor antigens such as Bcl-2, Bcl-xL, and surviving [81-84]. Another study examined the association between idarubicin and survivin expression in K562 cells of CML. The results showed that low-dose Idarubicin produced G2/MM arrest in CML cells, which was significantly related with increased survivin expression levels [70]. Furthermore, drug- resistant CML cell lines have been shown to respond to tuberostemonine by inhibiting survivin. The combination of tuberostemonine and doxorubicin has been shown to suppress cell proliferation and death in CML cells via downregulating survivin [85] (Table 2).
| Related cell lines | Correlation with surviving expression | References |
| P-gp in HL-60 cells | Does not appear to play a significant role as a IAP | 73-74 |
| Survivin in HL60R cells | Increased expression in HL-60R cells | 73-74 |
| NAIP in HL60R cells | Direct correlation with surviving expression | 73-74 |
| c-IAP2 in HL-60R cells | Direct correlation with surviving expression | 73-74 |
| Treatmentof HL60 cells with doxorubicin | Reduces X- IAP and survivin mRNA levels. | 74 |
| Treatmentof HL60 cells with cisplatin | Non-significant effect on survivin and NAIP mRNA levels. | 74 |
| Treatmentof HL60R cells with cisplatin | Survivin and NAIP are less influenced by the cisplatin therapy. | 74 |
| K562/ADR resistant cell line | This cell line has higher expression of surviving compared to K562 cells | 75 |
| imatinib-sensitive CML cells | This cell line has lower expression of survivin | 77-80 |
| Idarubicin relation with surviving expression in k562 cells | Survivin may promote apoptosis-resistant phenotype by inhibiting ida-induced apop-tosis. | 20 |
| Tuberostemonine in k562 cells | Inhibits surviving expression | 81 |
4-5 Survivin expression in leukemia
All of the individuals with adult T-cell leukemia (ATL) had high levels of survivin. However, the amounts of survivin mRNA in leukemic cells changed significantly between ATL subtypes, with low levels seen in chronic ATL cells and extremely high levels in acute ATL cells [86, 87]. Recently, Nakagawa et al. compared the expression of survivin in bone marrow cells from patients with acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia (CLL). Among 13 patients with ALL more than half the cases demonstrated positive expression of survivin while the majority of CLL cells exhibited intense expression of surviving [88]. ALL cells exhibited both nuclear and cytoplasmic survivin localization, whereas CLL cells mostly expressed survivin in the cytoplasm. Furthermore, there were no significant variations between survivin expression and the patient’s age, gender, or leukemic cell phenotype (B- or T-cell lineage) [88]. Some ALL samples showed high expression of other IAP family members such as XIAP, NAIP, and cIAP1. However, there was no significant difference in mean expression values across ALL patients and control groups. These findings may suggest that the high proliferative activity of ALL cells and the low proliferative state of CLL cells are connected with differing survivin localization patterns, and that survivin’s ability to prevent apoptosis is influenced by its placement inside the cell [88]. also The expression of cIAP2, XIAP and survivin was elevated together with caspase-3 and -7 inhibition after β1 integrin stimulation in pre-B ALL cells and integrin stimulation rescued pre-B cells from apoptosis [88]. XIAP was discovered to be expressed at varying levels in the majority of AML patients. The expression of cIAP1 and cIAP2 in AML has not been extensively explored.
In human, both cIAP1 and cIAP2 genes are localized on chromosome 11q22-q23, close to MLL gene, which is rearranged in acute leukemias with 11q23 aberration. Thus, their potential function as oncogenes in this subset of leukemias is to be elucidated [89]. The observations published so far may indicate a differential regulatory mechanism for the expression of IAP family proteins in different subtypes of leukemias.
4-6 Interaction between survivin protein and p53 tumor suppressor:
P53 (gene locus 17p13) is a nuclear phosphoprotein. It is a 20 kb tumor suppressor gene with 11 exons and ten introns. p53, a tumor suppressor gene, is located on human chromosome 17q23 and interferes with cell cycle regulation, DNA damage repair, and apoptosis. The p53 protein is a key regulator of the cell cycle. When p53 is activated, genes involved in cell cycle regulation at the G1 and G2 checkpoints, as well as apoptosis regulation, are induced or prevented from being expressed [85-92]. The p53 protein is activated in response to many cellular signals, including oncogene activation, which is implicated in cellular stress responses and causes cell cycle arrest, aging, or cell apoptosis. p53 stays at modest levels. When DNA was damaged, p53 became overexpressed [85-92]. Overexpression of p53 causes increased Bax expression and decreased Bcl2 expression, which is followed by caspase-9 activation and the intrinsic apoptosis pathway. p53 promotes p21-dependent cell cycle arrest or elevates pro-apoptotic Bcl-2 family proteins (BBax and Puma/ BBC3), resulting in apoptosis. p53 has been shown to reduce survival protein production through transcriptional regulation. An imbalance between pro-apoptotic proteins like p53 and anti-apoptotic proteins like Survivin causes tumor cells to become resistant to apoptosis [90-97]. p53 negatively regulates the expression of Survivin gene and they are regulators of cell and tissue homeostasis because of impress in the apoptotic pathways [90]. Survivin, also known as BIRC5 (gene locus: 17q25), is the smallest member of the inhibitor of apoptosis protein (IAP) gene family. It is one of the most frequently overexpressed genes in all forms of cancer and plays an important role in reducing apoptotic factors by limiting caspase activation. Caspase 3 and Caspase 7, for example, directly govern cell division in the G2 phase, as does Survivin. Survivin expression reduces cell death caused by multiple apoptotic events and regulates chromosome-microtubule adhesion. has proven that the increase in caspase-9 and expansion of the apoptotic signal leads to reduce the mRNA and protein expression of Survivin. [90-93, 95, 97-102]. Survivin has a high expression in the G2/M phase and rapidly drops in the G1 phase of the cell cycle, regulates cytokinesis progression, and participates in a range of pathways such as the p53 [63, 90]. Survivin expression, an unsatisfactory prognostic sign, correlates with decreased overall survival in different carcinomas [98].
Survivin gene expression is inhibited by p53; a balance between p53 and Survivin is required for cell viability [91, 94]. Survivin can also influence p53 expression, which regulates the p53-dependent apoptosis pathway [63]. P53 directly or indirectly binds to Survivin and inhibits it [103].
P53 uses a variety of variables to govern survival, including HDAC, p21, cdc25, HIF, DNMT1, E2F, and KLF5 (97). Survivin transcription is inhibited by p53 binding to the p53 binding site (p53BS) in the p53-Sp1 pathway, p53/EE2F pathway, and p53-dependent CDC25 regulation via CDE and CHR. CDE/CCHR regulates P21, PLK1, and cdc25; the p53-p21-RB/EE2F pathway regulates Survivin and cdc25; and the p53-p21-DREAM pathway regulates Survivin. DREAM includes DP, RB- like protein, E2F4, and MUVB [97]. Some research imply that p53 does not directly inhibit survivin. Survivin, on the other hand, is affected and suppressed by p53 via P21 and DREAM; DREAM binding sites such as CDE and CHR are employed to inhibit genes; Survivin is regulated by the p53-p21-DREAM-CDE/CCHR pathway; and Myc activates Survivin. The p53-p21-DREAM-CDE/CCHR pathway inhibits survivin, while MYC suppresses the p21 promoter and regulates the p53 transcriptional repression mechanism [102].
Another mediator is the transcription factor SP1, which remains in the Survivin promoter and works to attract more P53 to the Survivin promoter. P53, on the other hand, prevents HIF (hypoxia-inducing factor) from binding to the Birc5 promoter and thereby inhibiting survivin expression [104, 105].
p53 can also influence survival by activating DNMT1 (DNA methyltransferase 1). P53 recruits DNMT1 to enhance promoter hypermethylation, which is critical in epigenetic survival suppression [104-106].
P53 interacts with another factor known as histone lysine methyltransferase G9a. G9a catalyzes the H3K9 methylation process, resulting in dimethylated H3K9, which is an appropriate substrate for protein or HP1. The coordination of epigenetic regulators resulted in the creation of a chromatin-suppressor complex in the proximal area of the Survivin promoter, which eventually suppressed the Survivin gene [104].
A KLF5 transcription factor is involved in angiogenesis and cell proliferation. The interaction of KLF5 with the P53 tumor suppressor protein regulates apoptotic inhibitor proteins such as survivin. There are many GTboxes in the survivin promoter; KLF5 binds to the survivin promoter’s GT box sections, stimulating its activity; p53 can bind to KLF5, blocking KLF5 binding to the survivin promoter and reducing survivin expression. P53 leukemia patients are inactive, therefore KLF5 activates the survival promoter more strongly [107]
E2F is a factor that attaches to the survival promoter; P53 binds to E2F, forming the P53-E2F complex that inhibits survival gene expression [105] E2F is a factor that attaches to the survival promoter; P53 binds to E2F, forming the P53-E2F complex that inhibits survival gene expression [103].
P53 also binds to Sin3 and HDAC, leading to the formation of the P53-Sin3-HDAC complex and inhibiting Survivin [103], On the other side, p53 causes p21 expression, raises p21, decreases cdc2 expression, and subsequently leads to hypophosphorylation of pRB and its accumulation, which, coupled with E2F, suppresses Survivin expression. [103, 106]. However, some research imply that p53’s influence on Survivin is independent of p21 [106].
Therefore, p53 can influence Survivin expression by regulating various factors such as KLF5, E2f, SP1, Sin3, HDAC, DNMT1, p21 and CDC25 [101].
5- Survivin inhibition in cancers, especially leukemia
In recent years, many studies have focused on the use of survivin as a valuable therapeutic target in cancer treatment. Studies have shown that targeting survivors in human cancer enhances apoptosis and makes tumor cells more sensitive to chemotherapeutic treatments.
There are several strategies for targeting survivin, including the Ribozyme technique, immunotherapy, Dominant-negative mutant, and siRNAs [108].
5-1.Ribozyme trchnique
Ribozymes are small RNA molecules that are able to cleave the target RNA via endonucleolytic activity [109, 110]. Transfecting some ribozymes, such as RZ1 and RZ7, in human melanoma cell lines has been shown to lower survivin levels and increase caspase-9-dependent cell death. [110], indicating their anticancer function. However, none of these ribozymes have entered clinical trials [63]. In addition, two other ribozymes (RZ-1 and RZ-2) were demonstrated to be able to cleave at þ279 and þ289 nucleotides in human survivin mRNA [111], When cloned into an adenoviral vector, it boosted the etopside-induced apoptotic response in MCF-7 breast cancer cells [75]. Moreover, ribozymes bound to the ᶿ29 bacteriophage’s pRNA have been demonstrated in recent years as a potential remedy for ribozymes’ limited therapeutic potential [112].
5-2.Immunotherapy
The basis of cancer immunotherapy is the identification of specific tumor antigens. According to reports, CD8 + T cytotoxic cells exhibit high cytolytic activity against survivin epitopes. This research in animal models of pancreatic cancer and lymphoma that were responsive to survivin epitopes resulted in the development of survivin-based anticancer vaccines.
The vaccines were capable to induce tumor suppression in various cancers, including the pancreas [113], lung [114], neuroblastoma, and lymphoma [115]. Furthermore, the most significant advantage of these vaccinations is that no toxicity has been recorded thus far. The positive outcomes of these vaccine experiments have prompted researchers to focus on epitopes that potentially elicit the highest T-cell response against survivin. Some of these medications are now undergoing phase I and II clinical studies [109].
5-3.Dominant negative mutant
Gene therapy was one of the first strategies used to inhibit survivin activity. Initially, transfecting melanoma cells with survivin dominant-negative expression constructs was discovered to induce apoptosis [116]. Other investigations have shown that transfection with dominant-negative survivin mutants causes enhanced apoptosis in gastric cancer [117] and to repression of tumor growth in thymic lymphoma [118] and breast cancer [119] Animal models. Furthermore, survivin dominant-negative treatment has been demonstrated to improve tumor cells’ susceptibility to 5-fluorouracil and cisplatin [117]. Studies on mice expressing dominant-negative survivin revealed a reduction in tumor development and angiogenesis [117]. In addition, studies have represented that treating melanoma [120] and breast cancer [119, 121] xenografts with dominant-negative mutants of survivin repressed angiogenesis and increased apoptotic responses [119]. However, in vitro experiments revealed that these mutations had no influence on the proliferation of fibroblasts or normal endothelium cells [121].
5-3.other inhibitors
Small interfering RNAs (siRNAs) are double-stranded RNAs with 20-25 base pairs length that are able to inhibit gene expression. siRNAs are more successful and better therapeutic choices than other antisense techniques because they may be used at lower quantities, resulting in less side effects. The first siRNA against survivin was utilized in Hela cells, resulting in a delay in mitosis and prometaphase accumulation [122, 123]. Furthermore, in subsequent preclinical experiments, the use of siRNAs against survivin caused growth inhibition, increased apoptosis, and decreased survivin expression [123-126]. Furthermore, siRNA-mediated survivin repression boosted tumor cell susceptibility to a variety of therapies, including doxorubicin [127, 128], paclitaxel [129],
vincristine [130], TNF-alpha [127], and 17-allylamino- 17-demethoxygeldanamycin [131]. A research of mutant BRCA1 and knockdown BRCA1 cells found that raising survivin expression increased resistance to paclitaxel, whereas reducing survivin expression with siRNA restored cancer cell sensitivity to paclitaxel [132].
In conclusion, Survivin, a key anti-apoptotic protein, plays a critical role in leukemia by inhibiting programmed cell death and contributing to drug resistance. Its expression levels and localization vary across different leukemia subtypes, with notable differences observed between acute and chronic forms of the disease. Survivin’s interaction with tumor suppressor p53 further complicates its regulation, highlighting its importance in balancing cell survival and apoptosis.
Emerging therapeutic strategies targeting survivin— including ribozymes, immunotherapy, dominant-negative mutants, and small interfering RNAs—demonstrate potential for enhancing cancer treatment efficacy. By disrupting survivin’s function, these approaches aim to overcome resistance and improve patient outcomes, making survivin a promising target for future leukemia therapies.
Funding:
This research received no specific grant from any funding agency in the public, commercial, or not-for- profit sectors.
Acknowledgements
This study was supported by Tabriz university of medical sciences, Tabriz, Iran.
Conflict of interests
All the Authors declare no conflict of interests.
Data sharing statement
We hereby that data sharing is not applicable in our submission.
Ethical Approval
We hereby state that ethical approval is not applicable to our submission.
References
- The prevalence of oral manifestations in children with hematologic malignancys Pourshahidi S, et al . Elixir Journal.2012;49(8):9718-20.
- Hematopoietic modulators as potential agents for the treatment of leukemia Paredes-Gamero EJ , Nogueira-Pedro A, Miranda A, Justo GZ . Frontiers in Bioscience (Elite Edition).2013;5(1). CrossRef
- FOXM1 contributes to treatment failure in acute myeloid leukemia Khan I, Halasi M, Patel A, Schultz R, Kalakota N, Chen Y, Aardsma N, et al . JCI insight.2018;3(15). CrossRef
- Emerging Bone Marrow Microenvironment-Driven Mechanisms of Drug Resistance in Acute Myeloid Leukemia: Tangle or Chance? Ciciarello M, Corradi G, Forte D, Cavo M, Curti A. Cancers.2021;13(21). CrossRef
- Sex differences in adults with acute myeloid leukemia and the impact of sex on overall survival Stabellini N, Tomlinson B, Cullen J, Shanahan J, Waite K, Montero AJ , Barnholtz-Sloan JS , Hamerschlak N. Cancer Medicine.2023;12(6). CrossRef
- Acute myeloid leukemia in the real world: why population-based registries are needed Juliusson G, Lazarevic V, Hörstedt A, Hagberg O, Höglund M. Blood.2012;119(17). CrossRef
- Logroscino, Tumors in adolescents and young adults. 2016: Karger Medical and Scientific Publishers Beghi EG . .
- SEER cancer statistics review Ries LA , et al . 1975-2003. 2006..
- Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis Seif AE . Cancer Genetics.2011;204(5). CrossRef
- Age and acute myeloid leukemia Appelbaum FR , Gundacker H, Head DR , Slovak ML , Willman CL , Godwin JE , Anderson JE , Petersdorf SH . Blood.2006;107(9). CrossRef
- The influence of age on prognosis of de novo acute myeloid leukemia differs according to cytogenetic subgroups Schoch C, Kern W, Schnittger S, Büchner T, Hiddemann W, Haferlach T. Haematologica.2004;89(9). CrossRef
- Acute myeloid leukemia in children treated with epipodophyllotoxins for acute lymphoblastic leukemia Pui C. H., Ribeiro R. C., Hancock M. L., Rivera G. K., Evans W. E., Raimondi S. C., Head D. R., et al . The New England Journal of Medicine.1991;325(24). CrossRef
- Estimations of the increasing prevalence and plateau prevalence of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy Huang X, Cortes J, Kantarjian H. Cancer.2012;118(12). CrossRef
- Epidemiology of chronic myeloid leukaemia: an update Höglund M, Sandin F, Simonsson B. Annals of Hematology.2015;94 Suppl 2. CrossRef
- Stem cells, cancer, and cancer stem cells Reya T., Morrison S. J., Clarke M. F., Weissman I. L.. Nature.2001;414(6859). CrossRef
- Proliferation and apoptosis in acute and chronic leukemias and myelodysplastic syndrome Lin CW , Manshouri T, Jilani I, Neuberg D, Patel K, Kantarjian H, Andreeff M, et al , Albitar Maher, Estey Elihu. Leukemia Research.2002;26(6). CrossRef
- Targeting the apoptosis pathway in hematologic malignancies Zaman S, Wang R, Gandhi V. Leukemia & Lymphoma.2014;55(9). CrossRef
- Involvement of the bcl-2 gene in human follicular lymphoma Tsujimoto Y., Cossman J., Jaffe E., Croce C. M.. Science (New York, N.Y.).1985;228(4706). CrossRef
- Expression and prognostic significance of the inhibitor of apoptosis protein (IAP) family and its antagonists in chronic lymphocytic leukaemia Grzybowska-Izydorczyk O, Cebula B, Robak T, Smolewski P. European Journal of Cancer (Oxford, England: 1990).2010;46(4). CrossRef
- Survivin and leukemia Cong XI , Han ZC . International Journal of Hematology.2004;80(3). CrossRef
- Correlation between E-cadherin interactions, survivin expression, and apoptosis in MDCK and ts-Src MDCK cell culture models Capra J, Eskelinen S. Laboratory Investigation; a Journal of Technical Methods and Pathology.2017;97(12). CrossRef
- Down-regulation of survivin enhances paclitaxel-induced Hela cell apoptosis Gu F., Li L., Yuan Q.-F., Li C., Li Z.-H.. European Review for Medical and Pharmacological Sciences.2017;21(15).
- Validating survivin as a cancer therapeutic target Altieri DC . Nature Reviews. Cancer.2003;3(1). CrossRef
- 2016 Revision to the WHO Classification of Acute Myeloid Leukemia Hong M, He G. Journal of Translational Internal Medicine.2017;5(2). CrossRef
- Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death Joza N., Susin S. A., Daugas E., Stanford W. L., Cho S. K., Li C. Y., Sasaki T., et al . Nature.2001;410(6828). CrossRef
- Role of Smac in human leukaemic cell apoptosis and proliferation Jia L, Patwari Y, Kelsey SM , Srinivasula SM , Agrawal SG , Alnemri ES , Newland AC . Oncogene.2003;22(11). CrossRef
- Smac is required for cytochrome c-induced apoptosis in prostate cancer LNCaP cells Carson JP , Behnam M, Sutton JN , Du C, Wang X, Hunt , Weber MJ , Kulik G. Cancer Research.2002;62(1).
- An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif Crook N. E., Clem R. J., Miller L. K.. Journal of Virology.1993;67(4). CrossRef
- Targeting survivin in cancer Altieri DC . Cancer Letters.2013;332(2). CrossRef
- Caspase-independent cell death in AML: caspase inhibition in vitro with pan-caspase inhibitors or in vivo by XIAP or Survivin does not affect cell survival or prognosis Carter BZ , Kornblau SM , Tsao T, Wang R, Schober WD , Milella M, Sung H, Reed JC , Andreeff M. Blood.2003;102(12). CrossRef
- Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias Tamm I., Kornblau S. M., Segall H., Krajewski S., Welsh K., Kitada S., Scudiero D. A., et al . Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2000;6(5).
- Expression and prognostic significance of survivin in de novo acute myeloid leukaemia Adida C., Recher C., Raffoux E., Daniel M. T., Taksin A. L., Rousselot P., Sigaux F., et al . British Journal of Haematology.2000;111(1). CrossRef
- Expression of IAP family proteins in myelodysplastic syndromes transforming to overt leukemia Yamamoto K, Abe S, Nakagawa Y, Suzuki K, Hasegawa M, Inoue M, Kurata M, Hirokawa K, Kitagawa M. Leukemia Research.2004;28(11). CrossRef
- Induction of chemoresistance in HL-60 cells concomitantly causes a resistance to apoptosis and the synthesis of P-glycoprotein Campone M., Vavasseur F., Le Cabellec M. T., Meflah K., Vallette F. M., Oliver L.. Leukemia.2001;15(9). CrossRef
- XIAP expression is post-transcriptionally upregulated in childhood ALL and is associated with glucocorticoid response in T-cell ALL Hundsdoerfer P, Dietrich I, Schmelz K, Eckert C, Henze G. Pediatric Blood & Cancer.2010;55(2). CrossRef
- IAP inhibitors prime childhood leukemia cells to chemotherapy-induced apoptosis in a strictly RIP1-dependent manner and exert anti-leukemic activity in a NOD/SCID mouse model in vivo Löder S, et al . 2011, AACR..
- IAP proteins as targets for drug development in oncology Dubrez L, Berthelet J, Glorian V. OncoTargets and Therapy.2013;9. CrossRef
- Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies Smolewski P., Robak T.. Current Molecular Medicine.2011;11(8). CrossRef
- Targeting the IAP family of caspase inhibitors as an emerging therapeutic strategy Schimmer AD , Dalili S. Hematology. American Society of Hematology. Education Program.2005. CrossRef
- Inhibitor of Apoptosis (IAP) proteins as drug targets for the treatment of cancer Vaux DL . F1000 Biology Reports.2009;1. CrossRef
- Survivin: a unique target for tumor therapy Garg H, Suri P, Gupta JC , Talwar G. P., Dubey S. Cancer Cell International.2016;16. CrossRef
- Survivin does not influence the anti-apoptotic action of XIAP on caspase-9 Zumbrägel FK , Machtens DA , Curth U, Lüder CGK , Reubold TF , Eschenburg S. Biochemical and Biophysical Research Communications.2017;482(4). CrossRef
- Survivin at a glance Wheatley SP , Altieri DC . Journal of Cell Science.2019;132(7). CrossRef
- Survivin: A molecular biomarker in cancer Jaiswal PK , Goel A, Mittal R. D.. The Indian Journal of Medical Research.2015;141(4). CrossRef
- Study of the Expression Levels of Survivin Gene in Patients with Acute Myeloid Leukemia Abbassy HA , Elneely DAM , Rahman AA , Agram AESEAHA . International Journal For Research In Health Sciences And Nursing.2016;2(8). CrossRef
- Survivin is highly expressed in CD34(+)38(-) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML Carter BZ , Qiu Y, Huang X, Diao L, Zhang N, Coombes KR , Mak DH , et al . Blood.2012;120(1). CrossRef
- Inhibitor of apoptosis proteins in pediatric leukemia: molecular pathways and novel approaches to therapy Fulda S. Frontiers in Oncology.2014;4. CrossRef
- Expression of Survivin and Its Splice Variants in Pediatric Acute Lymphoblastic Leukemia Eren-Keleş E, Karabulut HG , Çakmaklı HF , Adaklı B, Köse SK , Uğur-Dinçaslan H, Yavuz G, Ertem M, Tükün A. Genetic Testing and Molecular Biomarkers.2018;22(12). CrossRef
- Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia Granziero L, Ghia P, Circosta P, Gottardi D, Strola G, Geuna M, Montagna L, et al . Blood.2001;97(9). CrossRef
- Survivin an important determinant for prognosis in adult T-cell leukemia: a novel biomarker in practical hemato-oncology Nakayama K, Kamihira S. Leukemia & Lymphoma.2002;43(12). CrossRef
- Cytokine-regulated expression of survivin in myeloid leukemia Carter B. Z., Milella M., Altieri D. C., Andreeff M.. Blood.2001;97(9). CrossRef
- The twisted survivin connection to angiogenesis Sanhueza C., Wehinger S., Castillo Bennett J., Valenzuela M., Owen G. I., Quest A. F. G.. Molecular Cancer.2015;14. CrossRef
- Analysis of the expression levels of survivin and VEGF in patients with acute lymphoblastic leukemia Yang M, Liu Y, Lu S, Wang Z, Wang R, Zi Y, Li J. Experimental and Therapeutic Medicine.2013;5(1). CrossRef
- Survivin structure: crystal unclear Shi Y.. Nature Structural Biology.2000;7(8). CrossRef
- Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement Verdecia M. A., Huang H., Dutil E., Kaiser D. A., Hunter T., Noel J. P.. Nature Structural Biology.2000;7(7). CrossRef
- Identification of a novel amplicon at distal 17q containing the BIRC5/SURVIVIN gene in malignant peripheral nerve sheath tumours Storlazzi C. T., Brekke H. R., Mandahl N., Brosjö O., Smeland S., Lothe R. A., Mertens F.. The Journal of Pathology.2006;209(4). CrossRef
- Enhancer element potentially involved in human survivin gene promoter regulation in lung cancer cell lines Mityaev M. V., Kopantzev E. P., Buzdin A. A., Vinogradova T. V., Sverdlov E. D.. Biochemistry. Biokhimiia.2010;75(2). CrossRef
- The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein Knauer SK , Bier C, Schlag P, Fritzmann J, Dietmaier W, Rödel F, Klein-Hitpass L, et al . Cell Cycle (Georgetown, Tex.).2007;6(12).
- Molecular analysis of survivin isoforms: evidence that alternatively spliced variants do not play a role in mitosis Noton EA , Colnaghi R, Tate S, Starck C, Carvalho A, Ko Ferrigno P, Wheatley SP . The Journal of Biological Chemistry.2006;281(2). CrossRef
- Nuclear survivin expression in mantle cell lymphoma is associated with cell proliferation and survival Martinez A, Bellosillo B, Bosch F, Ferrer A, Marcé S, Villamor N, Ott G, et al . The American Journal of Pathology.2004;164(2). CrossRef
- siRNA-mediated silencing of survivin inhibits proliferation and enhances etoposide chemosensitivity in acute myeloid leukemia cells Karami H, Baradaran B, Esfahani A, Estiar MA , Naghavi-Behzad M, Sakhinia M, Sakhinia E. Asian Pacific journal of cancer prevention: APJCP.2013;14(12). CrossRef
- Overexpression of survivin initiates hematologic malignancies in vivo Small S., Keerthivasan G., Huang Z., Gurbuxani S., Crispino J. D.. Leukemia.2010;24(11). CrossRef
- Survivin and Tumorigenesis: Molecular Mechanisms and Therapeutic Strategies Chen X, Duan N, Zhang C, Zhang W. Journal of Cancer.2016;7(3). CrossRef
- Role of the apoptotic and mitotic regulator survivin in melanoma McKenzie JA , Grossman D. Anticancer Research.2012;32(2). CrossRef
- Investigations of survivin: the past, present and future Cheung CHA , Cheng L, Chang K, Chen H, Chang J. Frontiers in Bioscience (Landmark Edition).2011;16(3). CrossRef
- Liaisons between survivin and Plk1 during cell division and cell death Colnaghi R, Wheatley SP . The Journal of Biological Chemistry.2010;285(29). CrossRef
- Role of Survivin in cytokinesis revealed by a separation-of-function allele Szafer-Glusman E, Fuller MT , Giansanti MG . Molecular Biology of the Cell.2011;22(20). CrossRef
- YM155 induces caspase-8 dependent apoptosis through downregulation of survivin and Mcl-1 in human leukemia cells Feng W, Yoshida A, Ueda T. Biochemical and Biophysical Research Communications.2013;435(1). CrossRef
- Study of survivin and X-linked inhibitor of apoptosis protein (XIAP) genes in acute myeloid leukemia (AML) Ibrahim AM , Mansour IM , Wilson MM , Mokhtar DA , Helal AM , Al Wakeel HM . Laboratory Hematology: Official Publication of the International Society for Laboratory Hematology.2012;18(1). CrossRef
- Survivin overexpression correlates with an apoptosis-resistant phenotype in chronic myeloid leukemia cells Nestal de Moraes G, Silva KL , Vasconcelos FDC , Maia RC . Oncology Reports.2011;25(6). CrossRef
- Developmental control of apoptosis by the immunophilin aryl hydrocarbon receptor-interacting protein (AIP) involves mitochondrial import of the survivin protein Kang BH , Xia F, Pop R, Dohi T, Socolovsky M, Altieri DC . The Journal of Biological Chemistry.2011;286(19). CrossRef
- Smac/DIABLO gene expression in acute myeloid leukemia patients Suliman GA , Mabrouk MM , Rabee ES , Gawaly A. The Egyptian Journal of Haematology.2013;38(2). CrossRef
- The role of the MDR1 (P-glycoprotein) gene in multidrug resistance in vitro and in vivo Roninson I. B.. Biochemical Pharmacology.1992;43(1). CrossRef
- Survivin mediates resistance to antiandrogen therapy in prostate cancer Zhang M, Latham DE , Delaney MA , Chakravarti A. Oncogene.2005;24(15). CrossRef
- A role for survivin in chemoresistance of endothelial cells mediated by VEGF Tran J, Master Z, Yu JL , Rak J, Dumont DL , Kerbel RS . Proceedings of the National Academy of Sciences of the United States of America.2002;99(7). CrossRef
- Changes in survivin messenger RNA level during chemotherapy treatment in ovarian cancer cells Wang Z, Xie Y, Wang H. Cancer Biology & Therapy.2005;4(7). CrossRef
- Expression of survivin messenger RNA correlates with poor prognosis in patients with hepatocellular carcinoma Ikeguchi M, Ueda T, Sakatani T, Hirooka Y, Kaibara N. Diagnostic Molecular Pathology: The American Journal of Surgical Pathology, Part B.2002;11(1). CrossRef
- Survivin transcription is associated with P-glycoprotein/MDR1 overexpression in the multidrug resistance of MCF-7 breast cancer cells Liu F, Liu S, He S, Xie Z, Zu X, Jiang Y. Oncology Reports.2010;23(5). CrossRef
- Resistance to diverse apoptotic triggers in multidrug resistant HL60 cells and its possible relationship to the expression of P-glycoprotein, Fas and of the novel anti-apoptosis factors IAP (inhibitory of apoptosis proteins) Notarbartolo M, Cervello M, Dusonchet L, Cusimano A, D'Alessandro N. Cancer Letters.2002;180(1). CrossRef
- Quantitative analysis of the anti-apoptotic gene survivin expression in malignant haematopoietic cells Moriai R., Asanuma K., Kobayashi D., Yajima T., Yagihashi A., Yamada M., Watanabe N.. Anticancer Research.2001;21(1B). CrossRef
- Regulation of survivin expression through Bcr-Abl/MAPK cascade: targeting survivin overcomes imatinib resistance and increases imatinib sensitivity in imatinib-responsive CML cells Carter BZ , Mak DH , Schober WD , Cabreira-Hansen M, Beran M, McQueen T, Chen W, Andreeff M. Blood.2006;107(4). CrossRef
- The antiapoptotic member of the Bcl-2 family Mcl-1 is a CTL target in cancer patients Andersen M. H., Becker J. C., Thor Straten P.. Leukemia.2005;19(3). CrossRef
- Spontaneous immunity against Bcl-xL in cancer patients Andersen MH , Reker S, Kvistborg P, Becker JC , Straten P. Journal of Immunology (Baltimore, Md.: 1950).2005;175(4). CrossRef
- BCR-ABL activity is critical for the immunogenicity of chronic myelogenous leukemia cells Brauer KM , Werth D, Schwarzenberg K, Bringmann A, Kanz L, Grünebach F, Brossart P. Cancer Research.2007;67(11). CrossRef
- Tuberostemonine reverses multidrug resistance in chronic myelogenous leukemia cells K562/ADR Wang YJ , Zhao HD , Zhu CF , Li J, Xie HJ , Chen YX . Journal of Cancer.2017;8(6). CrossRef
- Prognostic significance of survivin in pediatric acute lymphoblastic leukemia Esh AM , Atfy M, Azizi NA , El Naggar MM , Khalil EE , Sherief L. Indian Journal of Hematology & Blood Transfusion: An Official Journal of Indian Society of Hematology and Blood Transfusion.2011;27(1). CrossRef
- Adult T-cell leukemia cells overexpress Wnt5a and promote osteoclast differentiation Bellon M, Ko NL , Lee M, Yao Y, Waldmann TA , Trepel JB , Nicot C. Blood.2013;121(25). CrossRef
- Differential expression of survivin in bone marrow cells from patients with acute lymphocytic leukemia and chronic lymphocytic leukemia Nakagawa Y, Yamaguchi S, Hasegawa M, Nemoto T, Inoue M, Suzuki K, Hirokawa K, Kitagawa M. Leukemia Research.2004;28(5). CrossRef
- Expression of IAP-family proteins in adult acute mixed lineage leukemia (AMLL) Nakagawa Y, Hasegawa M, Kurata M, Yamamoto K, Abe S, Inoue M, Takemura T, et al . American Journal of Hematology.2005;78(3). CrossRef
- Deregulation of p53/survivin apoptotic markers correlated to PTEN expression in pterygium neoplastic cells Konstantopoulou K, Tsiambas E, Baliou E, Lazaris AC , Kavantzas N, Karameris A, Fotiades PP , et al . Journal of B.U.ON.: official journal of the Balkan Union of Oncology.2018;23(3). CrossRef
- Prognostic significance of survivin, β-catenin and p53 expression in urothelial carcinoma Senol S, Yildirim A, Ceyran B, Uruc F, Zemheri E, Ozkanli S, Akalin I, et al . Bosnian Journal of Basic Medical Sciences.2015;15(4). CrossRef
- Survivin protects fused cancer cells from cell death Do M, Kwak I, Ahn J, Lee I, Lee J. BMB reports.2017;50(7). CrossRef
- Berbamine induces SMMC-7721 cell apoptosis via upregulating p53, downregulating survivin expression and activating mitochondria signaling pathway Cao Y, Cao J, Yu B, Wang S, Liu L, Tao L, Sun W. Experimental and Therapeutic Medicine.2018;15(2). CrossRef
- Opposed arsenite-mediated regulation of p53-survivin is involved in neoplastic transformation, DNA damage, or apoptosis in human keratinocytes Li Y, Jiang R, Zhao Y, Xu Y, Ling M, Pang Y, Shen L, et al . Toxicology.2012;300(3). CrossRef
- Intensity of Nuclear Staining for Ki-67, p53 and Survivin as a New Prognostic Factor in Non-muscle Invasive Bladder Cancer Stec R, Cierniak S, Lubas A, Brzóskowska U, Syryło T, Zieliński H, Semeniuk-Wojtaś A. Pathology oncology research: POR.2020;26(2). CrossRef
- Pancreatic stellate cells increase pancreatic cancer cells invasion through the hepatocyte growth factor /c-Met/survivin regulated by P53/P21 Yang X, Liu S, Xu J, Cao S, Li Y, Zhou Y. Experimental Cell Research.2017;357(1). CrossRef
- CT‑1042, a novel anticancer agent, exhibits effects by activating p53 and inhibiting survivin Yang J, Li L, Xu C, Yang D, Wang S, Yuan S. Oncology Reports.2018;39(6). CrossRef
- YM155 Down-Regulates Survivin and Induces P53 Up-Regulated Modulator of Apoptosis (PUMA)-Dependent in Oral Squamous Cell Carcinoma Cells X Y, H S. Medical science monitor : international medical journal of experimental and clinical research.2017;23. CrossRef
- Integrin α5β1 and p53 convergent pathways in the control of anti-apoptotic proteins PEA-15 and survivin in high-grade glioma Renner G., Janouskova H., Noulet F., Koenig V., Guerin E., Bär S., Nuesch J., et al . Cell Death and Differentiation.2016;23(4). CrossRef
- Tumor expression of survivin, p53, cyclin D1, osteopontin and fibronectin in predicting the response to neo-adjuvant chemotherapy in children with advanced malignant peripheral nerve sheath tumor Karpinsky G, Krawczyk MA , Izycka-Swieszewska E, Fatyga A, Budka A, Balwierz W, Sobol G, et al . Journal of Cancer Research and Clinical Oncology.2018;144(3). CrossRef
- Selective inhibition of histone deacetylase 2 induces p53-dependent survivin downregulation through MDM2 proteasomal degradation Seo S, Hwang C, Choe T, Hong S, Yi J, Hwang S, Lee H, et al . Oncotarget.2015;6(28). CrossRef
- Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex Fischer M, Quaas M, Nickel A, Engeland K. Oncotarget.2015;6(39). CrossRef
- Structural, functional and therapeutic biology of survivin Sah N. K., Khan Z., Khan G. J., Bisen P. S.. Cancer Letters.2006;244(2). CrossRef
- Epigenetic mechanism of survivin dysregulation in human cancer Lyu H, Huang J, He Z, Liu B. Science China. Life Sciences.2018;61(7). CrossRef
- Survivin modulatory role in autoimmune and autoinflammatory diseases Pahlavan Y, Kahroba H, Samadi N, Karimi A, Ansarin K, Khabbazi A. Journal of Cellular Physiology.2019;234(11). CrossRef
- Survivin repression by p53, Rb and E2F2 in normal human melanocytes Raj D, Liu T, Samadashwily G, Li F, Grossman D. Carcinogenesis.2008;29(1). CrossRef
- KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia Zhu N, Gu L, Findley HW , Chen C, Dong J, Yang L, Zhou M. The Journal of Biological Chemistry.2006;281(21). CrossRef
- Targeting survivin in leukemia Carter BZ , Andreeff M. Oncology Reviews.2008;1(4). CrossRef
- Survivin as a preferential target for cancer therapy Mobahat M, Narendran A, Riabowol K. International Journal of Molecular Sciences.2014;15(2). CrossRef
- Molecular basis for the actions of Hsp90 inhibitors and cancer therapy Yamaki H, Nakajima M, Shimotohno KW , Tanaka N. The Journal of Antibiotics.2011;64(9). CrossRef
- Ribozyme-mediated cleavage of the human survivin mRNA and inhibition of antiapoptotic function of survivin in MCF-7 cells Choi KS , Lee TH , Jung MH . Cancer Gene Therapy.2003;10(2). CrossRef
- Survivin-directed Anticancer Therapies â A Review of Pre-clinical Data and Early-phase Clinical Trials Doolittle H, Morel A, Talbot D. 2010.
- Vaccination of hormone-refractory prostate cancer patients with peptide cocktail-loaded dendritic cells: results of a phase I clinical trial Fuessel S, Meye A, Schmitz M, Zastrow S, Linné C, Richter K, Löbel B, et al . The Prostate.2006;66(8). CrossRef
- A DNA vaccine targeting survivin combines apoptosis with suppression of angiogenesis in lung tumor eradication Xiang R, Mizutani N, Luo Y, Chiodoni C, Zhou H, Mizutani M, Ba Y, et al . Cancer Research.2005;65(2). CrossRef
- Survivin DNA vaccine generated specific antitumor effects in pancreatic carcinoma and lymphoma mouse models Zhu K, Qin H, Cha S, Neelapu SS , Overwijk W, Lizee GA , Abbruzzese JL , et al . Vaccine.2007;25(46). CrossRef
- Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma Grossman D., McNiff J. M., Li F., Altieri D. C.. The Journal of Investigative Dermatology.1999;113(6). CrossRef
- Suppression of survivin expression inhibits in vivo tumorigenicity and angiogenesis in gastric cancer Tu SP , Jiang XH , Lin MCM , Cui JT , Yang Y, Lum CT , Zou B, et al . Cancer Research.2003;63(22). CrossRef
- Effects of survivin antagonists on growth of established tumors and B7-1 immunogene therapy Kanwar J. R., Shen W. P., Kanwar R. K., Berg R. W., Krissansen G. W.. Journal of the National Cancer Institute.2001;93(20). CrossRef
- Cancer gene therapy using a survivin mutant adenovirus Mesri M., Wall N. R., Li J., Kim R. W., Altieri D. C.. The Journal of Clinical Investigation.2001;108(7). CrossRef
- Inhibition of melanoma tumor growth in vivo by survivin targeting Grossman D., Kim P. J., Schechner J. S., Altieri D. C.. Proceedings of the National Academy of Sciences of the United States of America.2001;98(2). CrossRef
- Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis Blanc-Brude OP , Mesri M, Wall NR , Plescia J, Dohi T, Altieri DC . Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2003;9(7).
- Survivin is required for stable checkpoint activation in taxol-treated HeLa cells Carvalho A, Carmena M, Sambade C, Earnshaw WC , Wheatley SP . Journal of Cell Science.2003;116(Pt 14). CrossRef
- Silencing of antiapoptotic survivin gene by multiple approaches of RNA interference technology Ling X, Li F. BioTechniques.2004;36(3). CrossRef
- Inhibition of survivin reduces cell proliferation and induces apoptosis in human endometrial cancer Ai Z, Yin L, Zhou X, Zhu Y, Zhu D, Yu Y, Feng Y. Cancer.2006;107(4). CrossRef
- Survivin downregulation by siRNA sensitizes human hepatoma cells to TRAIL-induced apoptosis Nakao K, Hamasaki K, Ichikawa T, Arima K, Eguchi K, Ishii N. Oncology Reports.2006;16(2).
- Survivin gene RNA interference inhibits proliferation, induces apoptosis, and enhances radiosensitivity in HeLa cells Song H, Xin X, Xiao F, Wang D, Yue Q, Han X. European Journal of Obstetrics, Gynecology, and Reproductive Biology.2008;136(1). CrossRef
- Transcriptional targeting of small interfering RNAs into cancer cells Huynh T, Wälchli S, Sioud M. Biochemical and Biophysical Research Communications.2006;350(4). CrossRef
- Small interfering RNA targeting survivin sensitizes lung cancer cell with mutant p53 to adriamycin Yonesaka K, Tamura K, Kurata T, Satoh T, Ikeda M, Fukuoka M, Nakagawa K. International Journal of Cancer.2006;118(4). CrossRef
- Regulation of microtubule stability and mitotic progression by survivin Giodini A, Kallio MJ , Wall NR , Gorbsky GJ , Tognin S, Marchisio PC , Symons M, Altieri DC . Cancer Research.2002;62(9).
- Lentivirus-mediated gene therapy by suppressing survivin in BALB/c nude mice bearing oral squamous cell carcinoma Jiang G, Li J, Zeng Z, Xian L. Cancer Biology & Therapy.2006;5(4). CrossRef
- Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells Paduano F, Villa R, Pennati M, Folini M, Binda M, Daidone MG , Zaffaroni N. Molecular Cancer Therapeutics.2006;5(1). CrossRef
- BRCA1 modulates malignant cell behavior, the expression of survivin and chemosensitivity in human breast cancer cells Promkan M, Liu G, Patmasiriwat P, Chakrabarty S. International Journal of Cancer.2009;125(12). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Nursing , 2025
Author Details
- Abstract viewed - 0 times
- PDF (FULL TEXT) downloaded - 0 times
- XML downloaded - 0 times