Involvement of p.R72P and PIN3 Ins16bp (TP53) Polymorphisms and the I157T (CHEK2) Mutation in Breast Cancer Occurrence in Burkina Faso
Download
Abstract
Introduction: The TP53 and CHEK2 genes have been described as breast cancer susceptibility genes and some of their polymorphisms have been associated with an increased risk of breast cancer in certain populations.
Aim: The objective of this study was to investigate the p.R72P and PIN3 Ins16bp (TP53) polymorphisms and the I157T (CHEK2) mutation developping of breast cancer.
Methods: This case-control study had enrolled 144 participants including 65 cases (breast cancer patients) and 79 controls (women without breast abnormalities) in the city of Ouagadougou in Burkina Faso. The DNA was extracted using the method of “salting out” and the genotyping of polymorphisms was performed by ASO-PCR (Allele Specific Oligonucleotides - Polymerase Chain Reaction), conventional PCR and PCR-RFLP (Polymerase Chain Reaction - Restriction Fragment Length Polymorphism) techniques.
Results: The heterozygous genotype (RP) of the p.R72P polymorphism of TP53 gene was in the majority in cases (73.85%) and controls (73.42%). Regarding to the PIN3 Ins16bp polymorphism of TP53 gene, the homozygous wild type (A1A1) was the most represented in both cases (53.85%) and controls (60.76%). Concerning the I157T mutation of CHEK2 gene, only one (01) patient was homozygous mutant (TT) and no controls had the mutation. This study found no association between these polymorphisms and the risk of breast cancer occurrence (p.R72P (OR=0.96; 95%IC (0.59-1.56); p=0.471), PIN3 Ins16bp (OR= 1.1; 95%IC (0.61-1.98); p=0.420)).
Conclusion: This study showed that the P allele of the p.R72P polymorphism and the wild-type allele (A1) of the PIN3 Ins16bp polymorphism were in the majority. The I157T mutation was very rare. These polymorphisms were not associated with the risk of developing breast cancer in this study.
Introduction
Breast cancer is a major public health problem. In 2020, 2.3 million new cases were diagnosed, representing 24.5% of all new cases of cancer in women worldwide [1]. In Burkina Faso, during 31 years, 14,587 cases of cancer have been diagnosed histologically in the anatomical pathology laboratories of Burkina Faso [2]. Among these cancers, breast cancer is the leading cancer in terms of incidence and prevalence It is also the most frequent cause of cancer mortality in women in Burkina Faso [2-4].
The exact causes of breast cancer are not clearly identified, but several studies have implicated a wide variety of factors in its genesis. Age, gender, heredity, reproductive factors, diet, the presence of other cancers, anthropometric characteristics, psychological and environmental factors as possible risk factors [5,6]. It’s reported that polymorphisms or mutations in some genes such as BRCA1, BRCA2, CHEK2, PALB2, TP53, ATM, PTEN are involved in the occurrence of breast cancer [7]. The TP53 gene is a tumor suppressor located on chromosome 17p13.1. It is 20kb of long and includes 11 exons and 10 introns [7]. It codes for a transcription factor: the p53 protein. The latter is a nuclear phosphoprotein with molecular weight of 53 kDa which is involved in the control of cell cycle progression, DNA repair, apoptosis and senescence [8-10]. Mutations or loss of function of the TP53 gene represent the most common genetic abnormality in sporadic breast cancer. It is altered in more than half of all human tumors, underscoring the importance of its function [11]. Its protein is considered as a “guardian of the genome” and plays many major roles in cell function [12].
Several polymorphisms of TP53 have been studied. The SNP p.R72P (rs1042522) of the TP53 gene is one of the most studied polymorphisms of TP53 gene [8,13]. It is located in exon and characterized by the presence of an arginine (R) or a proline (P) in position 72 [7]. This is explained by a substitution of a guanine (G) by a cytosine (C) at codon 72 of the TP53 gene [8]. This polymorphism exists in two isoforms (Arg72 and Pro72) which differ in their biochemical and biological properties [8]. Various meta-analysis studies have reported the involvement of p.R72P in the susceptibility to various cancer types, including gastric [14] and breast cancers [15]. However, studies on the association of breast cancer risk with the p.R72P polymorphism have not given consistent results [8]. The PIN3 Ins16bp polymorphism (rs17878362 is a duplication of 16 pairs of bases in intron 3 of TP53 gene. It affects mRNA splicing, modifying the coding regions [7]. This polymorphism was associated with reduced p53 mRNA levels and decreased apoptotic indices and DNA repair capacity in lymphoblastoid cell lines [16]. This explains its involvement in various cancers such as colon cancer [16] and breast cancer [17].
The CHEK2 gene is long of 50 kb and located on chromosome 22q12.1. Consisting of 14 exons, it codes for CHEK2, stable protein of 60 Da consisting consisting of 546 aa [18]. Activation of p53 by activated CHEK2 results in phosphorylation of p53, which stabilizes the p53 molecule [19,20]. Previous studies have shown that the association of CHEK2 with breast cancer is of low frequency and moderate penetrance in some countries [21]. The 1100delC and I157T mutations are among the most studied, conferring risks or not in the occurrence of breast cancer depending on the region and population studied. The I157T mutation is associated with an elevated risk of developing thyroid, breast, colon, kidney and prostate cancers [22]. Genetic predisposition to breast cancer in the african population is poorly studied [23].
The objective of this study is to determine the involvement of the p.R72P and PIN3 Ins16bp polymorphisms of the TP53 gene and the I157T of the CHEK2 gene in the development of breast cancer in a sample from Burkina Faso.
Materials and Methods
Site, type and period of study
This study was a case-control study conducted between October 2019 and October 2021 in the city of Ouagadougou, capital of Burkina Faso (West Africa). The study population consisted of 144 participants, including 65 patients with breast cancer histologically confirmed by a pathologist and 79 healthy controls without breast cancer who came for medical consultations at the University Hospitals: Yalgado Ouédraogo (CHU-YO) and Bogodogo (CHU-B); and at the Medical Centers with Surgical Branch (Paul VI and Schiphra). Biomolecular analyses were performed at the Laboratory of Molecular Biology and Genetics (LABIOGENE), and at the Pietro Annigoni Biomolecular Research Center (CERBA).
We are included in this study the cases related to all female patients with sporadic breast tumor or family history of breast cancer (hereditary) living in Burkina and whose the diagnosis is confirmed histologically by anatomo-pathologist. The cases were recruited during medical consultations (chemotherapy or chemotherapy interval check-up).
The controls included were all female living in Burkina, without any breast anomaly (as confirmed by mammography). Controls were selected from patients admitted to the same hospital for illnesses other than breast cancer.
We are excluded all men, any patient without histological confirmation of breast cancer ; any patient who voluntarily refuses to participate in the study.
Patients were classified as “sporadic breast cancer cases” if no one in their family had or currently has breast cancer. Patients were considered as with “family history of breast cancer” if at least one member of their family (niece, sister, mother, cousin, etc.) had or currently has breast cancer.
We are included all ages were in the study and all patients included in the study freely consented to their participation. We scrupulously respected confidentiality and anonymity. Written informed consents were obtained from each patient and control.
Our study was conducted according the Helsinki Declaration guidelines on ethics in medical research and obtained approval from the Health Research Ethics Committee (CERS) of Burkina Faso (Deliberation No. 2019-5-067 of 15 May 2019).
Sample collection and DNA extraction
The socio-demographic, anthropometric and residence data of the participants were obtained via questionnaires. The questionnaires were administered by trained interviewers or working in the field of breast cancer. According to patient’s residence, we used the classification of INSD [24]: urban if the women lived in town (province capital or urban municipality status) with a population greater than 10,000 inhabitants and rural if the community size was smaller. The age at diagnostic was determined by verification of identification card of each participant. Then, 5mLof venous blood from each consenting woman was collected on EDTA (Ethylene-Diamine-Tetra-Acetic) impregnated tubes, centrifuged at 13000 rpm for 15 min, plasma and pellet were stored at -20°C with individual codes. DNA was obtained from the blood pellets by the “Salting Out” technique as described by Miller et al. [25]. Then, using the Biodrop, the DNA extracts were quantified and the purity verified.
Molecular characterization of polymorphisms
The genotyping of the p.R72P and PIN3 Ins16bp polymorphisms of the TP53 gene was performed respectively by ASO-PCR and conventional PCR [26,7]. PCR-RFLP was used for the I157T mutation of the CHEK2 gene [27].
PIN3 Ins16bp polymorphisms of the TP53 gene and I157T of the CHEK2 gene
The reaction mix had a volume of 15μl for each polymorphism consisting 3μl of DNA, 2.8μl 5X FIREPol® Master Mix (Solis Biodyne), 8.2μl of sterile water, and 0.5μl of the R and F primers (0.5μM). The sequences of the primer pairs [7] are recorded in Table 1.
| Polymorphism | Primers | Size |
| p.R72P (rs1042522) of TP53 | Arg F: 5’-TCC CCC TTG CCG TCC CAA-3’ | Allele P: 177 bp |
| Arg R: 5’-CTG GTG CAG GGG CCA CGC-3’ | Allele R: 141 bp | |
| Pro F: 5’-GCC AGA GGT TGC TCC CCC-3’ | ||
| Pro R: 5’-CGT GCA AGT CAC AGA CTT-3’ | ||
| PIN3 Ins16bp (rs17878362) of TP53 | F: 5'-CTGAAAACA ACG TTC TGG TA-3 | Allele A1: 119pb |
| R: 5'-AAG GGG GAC TGT AGA TGG GTG-3' | Allele A2: 135pb | |
| I157T of CHEK2 | F: 5′- GCAAGAAACACTTTCGGATTTTCCGG -3′ | PCR: 194pb Digestion (PstI): |
| R: 5′- CCACTGTGATCTTCTATGTCTGCA-3′ | 20bp and 170pb | |
| β-globine | Forward: 5'-CAACTTCATCCACGTTCACC-3' | 260 bp |
| Reverse: 5'-GAAGAGCCAAGGACAGGTAC-3' |
The amplification was performed in a Gene Amp®PCR System 9700 (Applied Biosystems™) thermal cycler following the amplification programs: an initial denaturation at 95°C, 5 minutes, followed by 35 cycles of amplification (denaturation at 95°C for 45 sec, hybridization at 55°C for 30 sec (PIN3 Ins16bp of the TP53 gene), hybridization at 61.5°C for 40 sec (I157T of the CHEK2 gene), and elongation at 72°C for 45 sec) and a final extension at 72°C for 10 minutes. PCR products of the CHEK2 gene were digested with the restriction enzyme PstI [27]. In short, a reaction mix in a volume of 25μl consisting of 1μl of PstI enzyme (20U/μl), 5μl of Buffer 3.0 1X, 14μl of H2O, and 5μl of PCR product, was placed in each PCR plate. This mixture was then incubated at 37°C according to the manufacturer’s protocol for three (03) hours.
p.R72P polymorphism of the TP53 gene
The reaction mix had a total volume of 15μl for each polumorphism consisting of 3μl of DNA (10ng/μl), 2.8μl 5X FIREPol ®Master Mix (Solis Biodyne), 8.2μl of sterile water; 0.5μl of each primer pair of the R allele or P allele (0.5μM) and 0.5ul of the Reverse (R) and Forward (F) primers; (0.5μM) of the primers for β-globin gene have been used as internal control. The sequences of the primer pairs [28] are recorded in Table 1. The amplification was performed in a Gene Amp®PCR System 9700 (Applied Biosystems™) thermal cycler. The amplification program for the p.R72P polymorphism (R and P allele) was : an initial denaturation at 95°C, 15 minutes, followed by 35 cycles of amplification (denaturation at 95°C for 30 sec, hybridization at 60°C for 30 sec and elongation at 72°C for 30 sec) and a final extension at 72°C for 5 minutes. We have amplified the P allele and the R allele in separate reactions mix. For the internal control β-globin, the program was : an initial denaturation at 95°C, 15 minutes, followed by 10 cycles (β-globine) of amplification (denaturation at 95°C for 30 sec, hybridization at 60°C for 30 sec, and elongation at 72°C for 30 sec and a final extension at 72°C for 5 minutes.
PCR products were migrated on 2% gel for p.R72P and PIN3 Ins16bp of TP53 gene, 4% for I157T polymorphism of CHEK2 gene subjected to 100Volts and 80 mA voltage
Finally, DNA visualization was done with the Gene Flash Revelation (Synengege Bio Imaging, USA) instruments.
Validity of PCR amplication
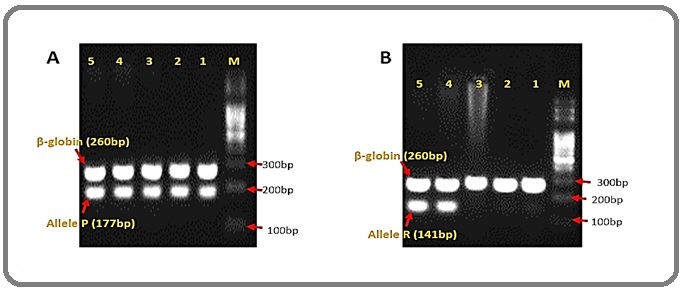
The validity of the PCR amplification of a sample for p.R72P polymorphism were conditioned by amplification of the β-globin gene (260bp) and it is invalid if no bands for β-globin were visible. The P allele is at 177bp and the R allele is at 141bp (Figure 1).
Figure 1. Gel Images of P.R72P of TP53 Gene. A, 1-5: allele P. B, 4-5: allele R; 1-3: no amplification allele R; M, Molecular weight marker; bp, base pairs.
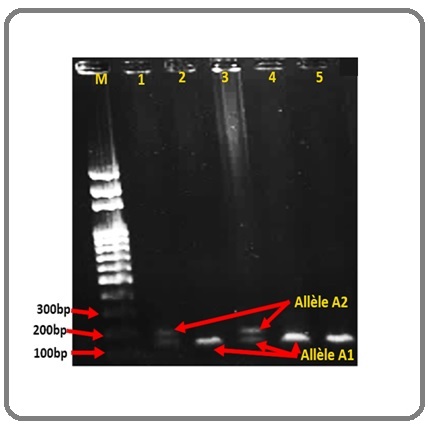
For PIN3 Ins16bp polymorphism of the TP53 gene, the presence of the alleles were conditioned by visualization of bands at 119bp for the A1 allele (wild type) and 135bp for the A2 allele (mutant) (Figure 2).
Figure 2. Gel Image of PIN3 Ins16bp; 1 and 3, Heterozygote A1A2; 2,4 and 5, Homozygote A1A1; M, Weight Marker; bp, Base Pairs.
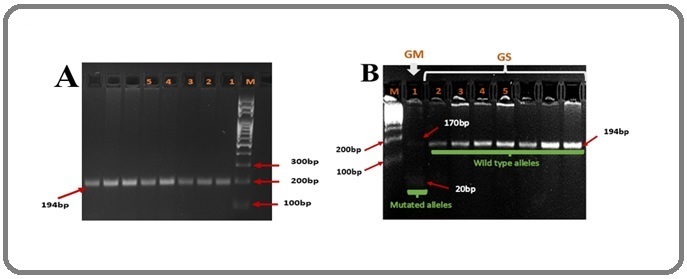
For I157T polymorphism of the CHEK2 gene, amplification is valid if the bands were observed at 194bp for the wild type allele (I) and 20bp/170bp for the mutant allele (T) (Figure 3).
Figure 3. Gel of I157T. A, Gel before Digestion, 194bp; B, Gel after Digestion; 1, Mutant Allele (T); 2-5, wild Type Allele (I); M, Molecular Weight Marker ; bp, Base Pairs ; GS, Wild Type; GM, Mutant Genotype.
Statistical analysis
The study data was entered into Excel. Epi Info version 7.1.1.14 (OpenEpi.com.) and Stata version 14.0 (Copyright 1985-2015 StataCorp LP, College Station, Texas 77845 USA) software were used for data analysis. Results were considered statistically significant for p -value ≤ 0.05.
The odds ratio (OR) and the 95% confidence intervals (CI) were calculated using the Epi Info version 7.1.1.14 (OpenEpi.com.) software to estimate associations between the carriage of alleles and certain sociodemographic parameters of breast cancer. The calculation was made by opposing two by two a variable to be studied with a reference variable.
Results
Sociodemographic characteristics of the study population
The study population consisted of 144 women, 65 of whom were cases (breast cancer patients) and 79 controls (non-breast cancer patients). The mean age of the cases was 44.51 ± 8.90 years and that of the controls 37.76 ± 11.01 years. Of all cases, 70.77% were older than 40 years. In addition, our results showed a significant difference between subjects over 40 years old and those under 40 years old (p < 0,001). This confirms previous studies that revealed that more than 40 years older have a higher risk than those with less than 40 years.
Also, among the patients, housewives were most represented (39.06%) followed by officials (35.94%) (Table 2).
| Subpopulation | Case | Controls | p -value |
| n (%) | n (%) | ||
| age range | |||
| >40 years | 46 (70.77) | 31 (39.74) | |
| ≤40 years | 19 (29.23) | 47 (60.26) | < 0.001 |
| Average age (years) | 44.51±8.9 | 37.76±11.01 | |
| BMI (kg/m²) | |||
| < 25 | 10 (22.73) | 19 (35.19) | |
| [25;30] | 10 (22.73) | 16 (29.63) | 0.151 |
| ≥30 | 24 (54.55) | 19 (35.19) | |
| Average BMI | 30.4±7.01 | 28.29±6.45 | - |
| Profession | |||
| Student | 2 (3.13) | 14 (17.95) | |
| Official | 23 (35.94) | 40 (51.28) | |
| Housewife | 25 (39.06) | 14 (17.95) | 0.002 |
| Particular | 13 (20.31) | 8 (10.26) | |
| Farmer | 1 (1.56) | 2 (2.56) | |
| Residence | |||
| Urban areas | 61 (93.85) | 79 (100) | |
| Rural areas | 4 (6.15) | 0 (0) | 0.08 |
The Body Mass Index (BMI) was calculated by taking the ratio of weight to height squared for each woman with these two parameters. According to the criteria of the “US National Institute of Health /National Heart Lung and Blood Institute (NCI/NHLBI)”, the indices were grouped into normal/lean (< 25 kg/m²); overweight (25 and 30 kg/m²) and obese (≥ 30 kg/m²). The patients had a mean BMI of 30.40±7.01 kg/m² and the controls 28.29±6.45 kg/m². Associations between BMI and disease did not show statistically significant results (p=0.151). (Table 2).
Clinical characteristics of the study population
In this study, parity and nulliparity didn’t show statistically significant difference (p>0.05) between cases and controls. Abortion, age at menarche, regularity of the menstrual cycle, age at first child were not also associated with a risk of breast cancer (Table 3). In the first hand, postmenopausal women are more exposed to breast cancer than those who are not yet (Table 3).
| Variables | Case | Controls | OR (95% CI) | p -value |
| Menopausal situation | ||||
| Not Post-Menopausal | 37 | 64 | Ref | |
| Menopaused | 28 | 15 | 3.23 (1.53-6.81) | 0.003 |
| Menstrual cycle | ||||
| Regular | 35 | 38 | Ref | |
| Irregular | 9 | 14 | 0.7 (0.27-1.81) | 0.617 |
| Abortion | ||||
| Nope | 37 | 54 | Ref | |
| Yes | 28 | 25 | 1.63 (0.83-3.23) | 0.214 |
| Age at menarche | ||||
| ≤ 12 years old | 4 | 10 | Ref | |
| > 12 years | 47 | 43 | 0.37 (0.11-1.25) | 0.174 |
| Parity | ||||
| Yes | 56 | 57 | Ref | |
| Nope | 9 | 22 | 0.42(0.18-0.98) | 0.067 |
| Age at first child | ||||
| ≤ 30 years old | 48 | 46 | Ref | |
| > 30 years | 8 | 9 | 0.85 (0.30-2.40) | 0.968 |
| Exogenous estrogen (contraception) | ||||
| Nope | 48 | 74 | Ref | |
| Yes | 17 | 5 | 5.24 (1.81-15.15) | 0.002 |
Furthermore, compared to those who did not used exogenous estrogens, the users were 5.24 times exposed to the risk of disease, and this difference is statistically significant (p=0.002). Indeed, all the exposed subjects, 77.27% were sick and 22.73% were free from the disease. Elsewhere in the unexposed subjects, 39.34% were affected by the disease and 60.66% were healthy. (Table 3).
In addition, the average number for parity in this study was 2.3 ± 1.8 children and the average age at first child was about 24 years (24.03 ± 0.61 years).
Allelic and genotypic frequencies
The P allele of the p.R72P polymorphism was the most prevalent in 50.77% of cases in our study population. The R allele was present in only 49.23% of the study participants. But, no association between the occurrence of breast cancer and the alleles R and P (OR=0.96; 95%IC (0.59-1.56); p=0.471) were found. The most present genotype of the p.R72P polymorphism was RP (heterozygous) with a proportion of 73.85% for cases and 73.42% for controls. The homozygous (RR) and (PP) genotypes were respectively 12.30% and 13.85% in the cases and 11.39% and 15.18% in the controls. Furthermore, no genotype was statistically associated (RR: OR=0.93; 95% IC (0.29-3.01); p=0.548 and PP: OR=0.84; 95%IC (0.19-3.68); p=0.527) with a risk of breast cancer (Table 4).
| Cases | Controls | OR (95 % IC) | P value | |
| N (%) | N (%) | |||
| Polymorphism p.R72P of TP53 gene | ||||
| Genotypes | ||||
| RR | 8 (12.31) | 9 (11.39) | - | Reference |
| RP | 48 (73.61) | 58 (73.42) | 0.93 (0.29-3.01) | 0.548 |
| PP | 9 (13.85) | 12 (15.19) | 0.84 (0.19-3.68) | 0.527 |
| Alleles | ||||
| R | 64 (49.23) | 76 (48.10) | - | Reference |
| P | 66 (50.77) | 82 (51.90) | 0.96 (0.59-1.56) | 0.471 |
| Genetic model of transmission of the mutated allele | ||||
| PP vs RP+ PP (Rec) | - | - | 0.9 (0.31-2.52) | 0.82 |
| PP+RP vs RR (Dom) | - | - | 0.92 (0.29-2.92) | 0.866 |
| Polymorphism PIN3 Ins16bp of TP53 gene | ||||
| Genotypes | ||||
| A1A1 | 35 (53.85) | 48 (60.76) | - | Reference |
| A1A2 | 29 (44.62) | 27 (34.18) | 1.47 (0.70-3.08) | 0.173 |
| A2A2 | 1 (1.54) | 4 (5.06) | 0.34 (0.01-3.7) | 0.315 |
| Alleles | ||||
| A1 | 99 (76.15) | 123 (77.85) | - | Reference |
| A2 | 31 (23.85) | 35 (22.15) | 1.1 (0.61-1.98) | 0.42 |
| Genetic model of transmission of the mutated allele | ||||
| A2A2 vs A1A2+ A2A2 (Rec) | - | - | 0.29 (0.01-3.08) | 0.25 |
| A2A2+A1A2 vsA1A1 (Dom) | - | - | 1.33 (0.65-2.72) | 0.403 |
| Polymorphism I157T of CHEK2 gene | ||||
| Genotypes | ||||
| II | 64 (98.46) | 79 (100) | - | Reference |
| IT | 0 (0) | 0 (0) | NA | - |
| TT | 1 (1.54) | 0 (0) | NA | - |
| Alleles | ||||
| I | 128 (98.46) | 158 (100) | - | Reference |
| T | 2 (1.54) | 0 (0) | NA | - |
| Genetic model of transmission of the mutated allele | ||||
| TT vs IT+ TT (Rec) | - | - | NA | - |
| TT+IT vs II (Dom) | - | - | NA | - |
Dom, dominant; Rec, recessive ; OR, Odd Ratio; 95%IC, 95% confidence interval
As for the PIN3 Ins16bp polymorphism, the most present allele was the wild type A1 with a proportion of 76.15%. The A2 mutant allele was present in only 23.85% of the study population. Furthermore, we did not find any risk associated with carrying both A1 and A2 alleles (OR= 1.1; 95%IC (0.61-1.98); p=0.420).
The A1A1 (homozygous wild type) genotype was the most present with a proportion of 53.85% in cases and 60.76% in controls. The A1A2 (heterozygous) and A2A2 (homozygous mutant) genotypes were respectively 44.62% and 1.54% for cases and 34.18% and 5.06% for controls. Here also, no genotype was statistically associated (A1A2: OR=1.47; 95%IC (0.70-3.08); p=0.173 and A2A2: OR=0.34 ;95%IC (0.01-3.7) t; p=0.315) with breast cancer risk in this study (Table 4).
Finally, for the I157T mutation, the mutant allele (T) was very rare (1.54%) in the cases and none control had the mutation. The wild-type allele (I) had a majority proportion of 98.46% in the cases and 100% in the controls. The most present genotype was the homozygous wild type (II) with a proportion of 98.46% in the cases and 100% in the controls in our study population. There were no heterozygous individuals (IT) in either the cases or controls. Only one (01) patient was homozygous mutant (TT). No controls had the mutation (Table 4).
Combined genotypes and the risk of developing breast cancer
We wanted to know if combined genotypes would increase the risk of developing breast cancer. Then, we paired the polymorphisms p.R72P and PIN3 Ins16bp of TP53. None of the combined polymorphism genotypes were associated with development (p>0.05) (Table 5).
| Genotypes | Controls | Cases | OR | 95%IC | P-value |
| N (%) | N (%) | ||||
| TP53 p.R72P and TP53 PIN3 Ins16bp | |||||
| RR-A1A1 | 9 (11,39) | 6 (9,23) | Ref | ||
| RR-A1A2 | - | 2 (3,08) | NA | NA | - |
| RP-A1A1 | 34 (43,04) | 25 (38,46) | 1.1 | 0.35-3.5 | 0.9 |
| RP-A1A2 | 20 (25,32) | 23 (35,38) | 1.73 | 0.52-5.69 | 0.549 |
| RP-A2A2 | 4 (5,06) | - | NA | NA | - |
| PP-A1A1 | 5 (6,33) | 4 (6,15) | 1.2 | 0.23-6.39 | 0.831 |
| PP-A1A2 | 7 (8,86) | 4 (6,15) | 0.86 | 0.17-4.27 | 0.826 |
| PP-A2A2 | - | 1 (1,54) | NA | NA | - |
NA, not applicable; OR, Odds Ratio; 95%CI, 95% confidence interval
Discussion
Sociodemographic data
The mean age of patients in our study population was 44.51±8.9 years which is lower than those found by Bambara et al., (2017) [3] which was 48.20 ± 12.4 years and by Zongo et al., (2021) [2] which was 47.5 years in Burkina Faso. The mean age of patients seems to decrease over the years.
In this study, only 6.15% of the patients lived in rural areas. A study in Nigeria showed that urban dwellers who were in daily contact with exhaust fumes and industrial waste were at higher risk of breast cancer than those who were not (OR=6.91; p=0.00), including rural dwellers [28]. In addition, Ba et al., (2020) [29] showed the difficulties of rural women to consult a health practitioner for their health problems in sub-Saharan Africa. All of these observations could explain the trend of high breast cancer prevalence in African cities compared to low rates of breast cancer in rural areas.
In this study, the association between BMI and disease did not show a significant difference (p=0.151) but we found that the mean BMI of the patients was slightly higher (30.4±7.01 kg/m²) than that of the controls (28.29±6.45 kg/m²). Some previous studies have associated obesity with an increased risk of breast cancer [30], other studies haven’t found obesity to be associated with a risk of breast cancer in Burkina Faso [31].
Reproductive Factors
The average number for parity in this study is 2.3 ± 1.8 children and the mean age at first birth was about 24 years (24.03 ± 0.61 years). In the past, African women were characterized by an advanced age at menarche (after 15 years); the birth of the first child at a relatively early age (around 19 years old) and by a relatively high multiparity (5 to 9 children per woman), which would protect them from breast cancer [32]. But the adoption of Western lifestyles nowadays contributes to the regression of these protective factors among African women [33]. This last observation is a reality in our study population.
In our study, associations between nulliparity and multiparity were not associated with the disease occurrence (p=0.067). On the other hand, multiparity is often associated with protection and nulliparity with a risk of breast cancer. Full-term pregnancy and breastfeeding have been shown to contribute to a decrease in estrogen levels and increases this protection against the development of breast cancer [34].
Our results do not indicate a significant difference (p=0.174) between the age at menarche, the irregularity of the menstrual cycle and the development of breast cancer. Our results confirms that of a nigerian study [35] who found no association between age at menarche; parity and risk of developing breast cancer in a study of a cohort of 2000 Nigerian participants. But other studies have reported that an early menarche before the age of 12 increases the risk of developing breast cancer [36].
We also associated the menopausal situation with the disease. Menopause would increase the risk of developing the disease (p=0.002). Among postmenopausal women, patients were 65.12% versus 34.88% for controls and among women who had not yet reached menopausal age, patients were 36.63% versus 63.37 % for controls. This result confirms what Sun et al., reported in 2017 that the risk of breast cancer increases by 3% each year after menopause [37]. But other studies did not find a direct association between breast cancer and age at menopause [38].
Allelic and genotypic frequencies of the p.R72P, PIN3 Ins16bp and I157T polymorphisms
Our results show that the P allele of the p.R72P polymorphism was the most frequent in both cases (50.77%) and controls (51.90%) and the R allele had a proportion of 49.34% and 48.10% in cases and controls respectively but no association were found between an allele of this polymorphism and breast cancer. In Tunisian population, a study [39] have not found also association between an allele of the R72P polymorphism and breast cancer but it have showed that the R allele was the frequent in both cases (75%) and controls (65.3%). Out of Africa, our frequencies are slightly different from those obtained by a study in an Indian population [7] where the R allele was most frequent in controls (53.8%) and the P allele most frequent in cases (56.9%). Here also, none allele were associated with brest cancer. In addition, the P allele has been associated with the risk of developing colorectal cancer in the Japanese [40], Korean [41], Chinese [42] and Malaysian [43] population while the R allele has been associated with the risk of developing breast cancer in Greece [44]. It is also reported that the R allele induces apoptosis more efficiently than the P allele [8,45,46]. On the other hand, the P variant is more effective in inducing cell cycle arrest in the G1 phase, allowing better repair of damaged DNA [8,47].
At the genotypic level, the heterozygous genotype (RP) was in the majority in both cases (73.85%) and controls (73.41%) but none genotype were associated with breast cancer. These results are different to those of Guleria et al (2012) [7] in India where heterozygotes were 58.8% in cases and 40% in controls; no association were also reported. Furthermore, the association of PP genotype and increased risk of breast [48], colorectal [49], bladder cancer in the North Indian population have been reported. However, no association was found between breast cancer and the p.R72P polymorphism in Tunisian [39] and Russian [50] patients.
As for the PIN3 Ins16bp polymorphism, the A1 (wild type) allele was the most frequent in both cases (76.15%) and controls (77.85%) and the A2 (mutant) allele had a proportion of 23.85% and 22.15% respectively in cases and controls. We have not found a study in Africa on this polymorphism but these frequencies are similar to those obtained in a study in an Indian population [7] where the A1 allele was the most frequent in both patients (72.5%) and controls (81.9%). The A2 allele had a proportion of 27.5% and 18.1% in cases and controls respectively. At the genotypic level, the wild type (A1A1) was the majority in both cases (53.85%) and controls (60.76%). These results are similar to those of Guleria et al. (2012) in India where wild-type genotypes were the most frequent (53.8% in cases and 66.3% in controls) [7]. The A2A2 genotype has been reported as a risk for the development of breast cancer [51] . Also a high risk of A1A2 genotype in the development of breast cancer has been reported in Iranian population [52]. It is reported that the PIN3 Ins16bp polymorphism is a duplication of 16 base pairs in intron 3 of TP53 gene. It affects mRNA splicing, altering coding regions [7]. The involvement of this polymorphism in cancers such as colon cancer [16] and breast [17] is a consequence of its association to the reduction of p53 mRNA level and decreased apoptotic indices as well as DNA repair capacity in lymphoblastoid cell lines [16].
Finally, for the I157T mutation of the CHEK2 gene, it was found in only one patient in our population. Similarly, the I157T mutation has not been identified in different populations [27,53,54] . Moreover, this mutation has not been associated with an increased risk in the Moroccan population [55]. On the other hand, a large study [56] reported the c.1343T>G mutation of CHEK2 in African subjects. This mutation was associated with a risk of prostate cancer in African men. In this same study three (03) mutations of CHEK2 gene including c.349A> G; c.1036C>T; c.538C>T was associated with breast cancer risk in European women and the c.1312G>T mutation was associated with prostate cancer risk in European men [56]. We did not find an African study reporting the presence of this mutation in Africa. It is reported that following a break in the double-stranded DNA via the action of an ionizing agent or a genotoxic substance, CHEK2 will be activated by phosphorylation and homodimerization by ATM kinase. After the activation of CHEK2, this molecule will activate other molecules such as p53, BRCA1, Cdc25A and Cdc25C. Activation of p53 by activated CHEK2 occurs through phosphorylation of p53 which stabilizes the p53 molecule [19,20]. All this results in consequences such as cell arrest in G1, S and G2/M or activation of DNA repair or apoptosis in certain cases. Thus, the I157T mutation of the CHEK2 gene produces a protein that is stable but unable to recognize p53, Cdc25 and BRCA1. This mutation takes place at the level of exon 3 [57] and leads to the formation of tumors by a dominant negative effect rather than by dysfunction [20].
In this study, none of these polymorphisms have been directly associated with the risk of developing breast cancer. Thus, the p.R72P and PIN3 Ins16bp polymorphisms of the TP53 gene as well as the I157T mutation of the CHEK2 gene do not appear to be genetic factors involved in the development of breast cancer in our study population. But muticenter studies and on large samples are needed before extending this result at national level.
Associated genotypes
Generally, the effect of a single SNP is weak to implicate a pathology but the combined effects of aberrant SNPs may contribute additively or synergistically to a risk of developing a pathology. In our study, the polymorphisms p.R72P, PIN3 Ins16bp and I157T associated two by two in correlation with the development of breast cancer did not show significant differences (p>0.05). We have not found previous studies regarding the association between p.R72P, PIN3 Ins16bp and I157T polymorphisms in breast cancer. The no association between the combined genotypes and the deseases can be justified by the relatively small sample size that influences the statistical power.
Limits of this study
Despite all the efforts made, this study had some limits as the reduced size of the sample. Also, some clinicopathological parameters were not found in the medical records of patients, which weakened the multivariate analysis. The ages between patients and controls were not matched. Also, the subjects of this studies came from hospitals in the only city of Ouagadougou. For future investigations, several cities must be included in the study.
Perspectives and Challenges
We have in prospects to extend this study to several cities in Burkina Faso to clearly establish the involvement of these polymorphism in development of breast cancer at national level. In addition does not there be a way to restore the alterations of TP53 gene of sporadic cancers? This is a question that we will try to answer in our future studies.
In conclusion, this study showed that the allele P of the p.R72P polymorphism and the wild-type allele (A1) of the PIN3 Ins16bp polymorphism were in the majority. As for the I157T mutation, it was very rare in our study population. In short, in our study, none of these polymorphisms were associated with the risk of developing breast cancer, so they do not appear to be genetic factors involved in the development of breast cancer in our study population. However, given the small size of our population, other investigations of the TP53 and CHEK2 genes in a large population may help to provide definitive informations.
Acknowledgements
The authors sincerely acknowledge all the participants (cases and controls), Laboratory of Molecular and Genetic Biology (LABIOGENE) and Pietro Annigoni Biomolecular Research Center (CERBA), for their valuable contribution to the successful completion of the study.
Statement
This manuscript has been published as a preprint according to the following link: https://www. researchsquare.com/article/rs-1693953/v1 ORCID link of the corresponding author: https://orcid. org/0000-0002-9423-024X.
Ethics approval and consent to participate
Our study obtained approval from the Health Research Ethics Committee (CERS) of Burkina Faso (Deliberation No. 2019-5-067 of 15 May 2019). All participants gave their free and informed consent. We scrupulously respected confidentiality and anonymity. A written consent were obtained from every patient and control.
Availability of data and material
The data used in this study are available and will be provided by the corresponding author on a reasonable request.
Competing interests
The authors declare that they have no conflict of interest.
Funding statement
No funding announcement.
Authors’ contributions
Conceptualization, JS, AAZ and SD; Methodology: SD, AAZ, TIK, LJA, HKS, MDWA, BSB, LT, TCO and RAO; Formal analysis and investigation: SD, AAZ, HKS, TIK, MDWA and LT; Writing-original draft preparation: AAZ and SD; Writing-review and editing: TIK, LJA,HKS, MDWA, BSB, LT, TCO, RAO, NZ, TMZ, ATY, AYS and FWD; patient enrolment: NZ, AYS, ITK and LJA. Supervision: ATY and JS.
All authors were the major contributors in writing the manuscript. All authors read and approved the final manuscript.
References
- GLOBOCAN [Internet]. GLOBOCAN, 20202020 [cité 2021 févr 23];Available from: http://gco.iarc.fr/today/home .
- Incidences et évolution des fréquences des cancers au Burkina Faso de 1988 à 2018 Zongo N, Ouédraogo A, Ouédraogo S, Sanon-Lompo S, Ido FA , Djiguemdé A, et al . BURKINA MEDICAL 2021;25:15..
- Breast cancer: descriptive profile of 80 women attending breast cancer care in the Department of General and Digestive Surgery of CHU-YO Bambara HA , Zouré AA , Sawadogo AY , Ouattara AK , Marie NL , Traoré SS , et al . Pan Afr Med J [Internet] 2017 [cité 2021 juin 21];28. Available from: http://www.panafrican-med-journal.com/content/article/28/314/full/..
- Ministere De La Sante Burkina Faso. Plan Strategique De Lutte Contre Le Cancer 2013 - 2017. 2013;74 .
- Epidemiology of breast cancer Key TJ , Verkasalo PK , Banks E. The Lancet. Oncology.2001;2(3). CrossRef
- Epidemiology of breast cancer in Malaysia Yip CH Cheng Har, Taib NAM Nur Aishah Mohd, Mohamed I. Asian Pacific journal of cancer prevention: APJCP.2006;7(3).
- p.R72P, PIN3 Ins16bp polymorphisms of TP53 and CCR5?32 in north Indian breast cancer patients Guleria K, Sharma S, Manjari M, Uppal MS , Singh NR , Sambyal V. Asian Pacific journal of cancer prevention: APJCP.2012;13(7). CrossRef
- TP53 polymorphisms in sporadic North Indian breast cancer patients Sharma S, Sambyal V, Guleria K, Manjari M, Sudan M, Uppal MS , Singh NR , Bansal D, Gupta A. Asian Pacific journal of cancer prevention: APJCP.2014;15(16). CrossRef
- [Exquisite sensitivity of TP53 mutant breast cancers to dose-dense chemotherapy] Lehmann-Che J, Turpin E, Bertheau P, Espié M, Thé H. Medecine Sciences: M/S.2007;23(11). CrossRef
- The first 30 years of p53: growing ever more complex Levine AJ , Oren M M. Nature Reviews. Cancer.2009;9(10). CrossRef
- [Generalities about carcinogenesis] Tubiana M. Comptes Rendus Biologies.2008;331(2). CrossRef
- [Genetic bases of the radiosensitivity of breast cancer] Delaloge S, Marsiglia H. Cancer Radiotherapie: Journal De La Societe Francaise De Radiotherapie Oncologique.2005;9(2). CrossRef
- p53 mutation heterogeneity in cancer Soussi T, Lozano G. Biochemical and Biophysical Research Communications.2005;331(3). CrossRef
- P53 codon 72 polymorphism and gastric cancer: a meta-analysis of the literature Zhou Y, Li N, Zhuang W, Liu GJ , Wu TX , Yao X, Du L, Wei ML , Wu XT . International Journal of Cancer.2007;121(7). CrossRef
- P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case-control studies Zhang Z, Wang M, Wu D, Wang M, Tong N, Tian Y, Zhang Z. Breast Cancer Research and Treatment.2010;120(2). CrossRef
- A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutiérrez-Enríquez S, Hall J, Guino E, Peinado MA , Capella G, Canzian F. Oncogene.2004;23(10). CrossRef
- Importance of TP53 codon 72 and intron 3 duplication 16bp polymorphisms in prediction of susceptibility on breast cancer Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, Schmitt F. BMC cancer.2008;8. CrossRef
- Analysis of CHEK2 FHA domain in Czech patients with sporadic breast cancer revealed distinct rare genetic alterations Kleibl Z, Havranek O, Novotny J, Kleiblova P, Soucek P, Pohlreich P. Breast Cancer Research and Treatment.2008;112(1). CrossRef
- CHK2 kinase--a busy messenger Bartek J, Falck J, Lukas J. Nature Reviews. Molecular Cell Biology.2001;2(12). CrossRef
- Chk1 and Chk2 kinases in checkpoint control and cancer Bartek J, Lukas J. Cancer Cell.2003;3(5). CrossRef
- Gene analysis techniques and susceptibility gene discovery in non-BRCA1/BRCA2 familial breast cancer Aloraifi F, Boland MR , Green AJ , Geraghty JG . Surgical Oncology.2015;24(2). CrossRef
- CHEK2 is a multiorgan cancer susceptibility gene Cybulski C., Górski B., Huzarski T., Masojć B., Mierzejewski M., Debniak T., Teodorczyk U., Byrski T., Gronwald J., Matyjasik J., Zlowocka E., Lenner M., Grabowska E., Nej K., Castaneda J., Medrek K., Szymańska A., Szymańska J., Kurzawski G., Suchy J., Oszurek O., Witek A., Narod S. A., Lubiński J.. American Journal of Human Genetics.2004;75(6). CrossRef
- The TP63 Gene Polymorphism rs17506395 is Associated with Early Breast Cancer in Cameroon Tiofack Arnol T. Z., Simo Gustave, Ofon Elvis, Dina-Bell Esther, Kamla Chancelin M., Ananga Sidonie N., Roger Tchamfong, Nana Theophile N., Ngeufack Charlotte T., Fewou Adamou, Takongmo Samuel, Lueong Smiths. Asian Pacific journal of cancer prevention: APJCP.2020;21(8). CrossRef
- Institut national de la statistique et de la démographie (INSD). Annuaire statistique [Internet]. Burkina Faso: 2014 Available from: http://www.insd.bf/contenu/pub_periodiques/annuaires_stat/Annuaires_stat_nationaux_BF/Annuaire_stat_2014.pdf..
- A simple salting out procedure for extracting DNA from human nucleated cells. Miller SA, Dykes DD, Polesky HF. Nucleic Acids Research.1988;16(3).
- Polymorphism of TP53 codon 72 showed no association with breast cancer in Iranian women Khadang B, Fattahi MJ , Talei A, Dehaghani AS , Ghaderi A. Cancer Genetics and Cytogenetics.2007;173(1). CrossRef
- An association study between CHEK2 gene mutations and susceptibility to breast cancer Jalilvand M, Oloomi M, Najafipour R, Alizadeh SA , Saki N, Rad FS , Shekari M. Comparative Clinical Pathology.2017;26(4). CrossRef
- Risk Factors Associated with Breast Cancer among Women in Warri and lbadan, Nigeria Oladimeji KE , Ajayi IO , Okareh OT . Nigerian Health Journal.2013;13(3).
- Prevalence and determinants of breast cancer screening in four sub-Saharan African countries: a population-based study Ba DM , Ssentongo P, Agbese E, Yang Y, Cisse R, Diakite B, Traore CB Cheick Bougadari, et al . BMJ open.2020;10(10). CrossRef
- Risk factors for fatal breast cancer in African-American women and White women in a large US prospective cohort McCullough ML , Feigelson HS , Diver WR , Patel AV , Thun MJ , Calle EE . American Journal of Epidemiology.2005;162(8). CrossRef
- Carriage of HLA-DRB1*11 and 1*12 alleles and risk factors in patients with breast cancer in Burkina Faso Zouré AA , Amegnona LJ , Zongo N, Kiendrebeogo IT , Sorgho PA , Zongo FI , Yonli AT , et al . Open Life Sciences.2021;16(1). CrossRef
- Breast cancer in sub-Saharan Africa: how does it relate to breast cancer in African-American women? Fregene A, Newman LA . Cancer.2005;103(8). CrossRef
- Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries Sung H, Ferlay J, Siegel RL , Laversanne M, Soerjomataram I, Jemal A, Bray F. CA: a cancer journal for clinicians.2021;71(3). CrossRef
- [Risk factors and risk reduction of breast cancer] Nkondjock A, Ghadirian P. Medecine Sciences: M/S.2005;21(2). CrossRef
- Breast cancer in Nigerian women Ihekwaba FN. The British Journal of Surgery.1992;79(8). CrossRef
- Breast cancer in Saudi Arabia and its possible risk factors. Almutlaq BA , Almuazzi RF , Almuhayfir AA , Alfouzan AM , Alshammari BT , AlAnzi HS , et al . Journal of Cancer Policy [Internet] 2017 [cité 2020 déc 29];12:83‑9. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2213538316300558..
- Risk Factors and Preventions of Breast Cancer Sun YS , Zhao Z, Yang ZN , Xu F, Lu HJ , Zhu ZY , Shi W, Jiang J, Yao PP , Zhu HP . International Journal of Biological Sciences.2017;13(11). CrossRef
- Case-control study of risk factors for breast cancer in Nigerian women Okobia M, Bunker C, Zmuda J, Kammerer C, Vogel V, Uche E, Anyanwu S, Ezeome E, Ferrell R, Kuller L. International Journal of Cancer.2006;119(9). CrossRef
- No evidence of correlation between p53 codon 72 polymorphism and risk of bladder or breast carcinoma in Tunisian patients Mabrouk I, Baccouche S, El-Abed R, Mokdad-Gargouri R, Mosbah A, Saïd S, Daoud J, et al . Annals of the New York Academy of Sciences.2003;1010. CrossRef
- Allele Frequencies of 25 Polymorphisms Pertaining to Cancer Risk for Japanese, Koreans and Chinese Hamajima N, Takezaki T, Tajima K. Asian Pacific journal of cancer prevention: APJCP.2002;3(3).
- The p53 codon 72 polymorphism and susceptibility to colorectal cancer in Korean patients Cao Z, Song JH , Park YK , Maeng EJ , Nam SW , Lee JY , Park WS . Neoplasma.2009;56(2). CrossRef
- Association of the TP53 codon 72 polymorphism with colorectal cancer in a Chinese population Zhu ZZ , Wang AZ , Jia HR , Jin XX , He XL , Hou LF , Zhu G . Japanese Journal of Clinical Oncology.2007;37(5). CrossRef
- Association of Arg72Pro of P53 polymorphism with colorectal cancer susceptibility risk in Malaysian population Aizat AAA , Shahpudin SNM , Mustapha MA , Zakaria Z, Sidek ASM , Abu Hassan MR , Ankathil R. Asian Pacific journal of cancer prevention: APJCP.2011;12(11).
- p53 codon 72 polymorphism as a risk factor in the development of breast cancer Papadakis EN, Dokianakis DN, Spandidos DA. Molecular cell biology research communications: MCBRC.2000;3(6). CrossRef
- The codon 72 polymorphic variants of p53 have markedly different apoptotic potential Dumont P, Leu JL , Della Pietra AC , George DL , Murphy M. Nature Genetics.2003;33(3). CrossRef
- p53 polymorphic variants at codon 72 exert different effects on cell cycle progression Pim D, Banks L. International Journal of Cancer.2004;108(2). CrossRef
- Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population Ørsted DD , Bojesen SE , Tybjaerg-Hansen A, Nordestgaard BG . The Journal of Experimental Medicine.2007;204(6). CrossRef
- A Case-Control Study of TP53 R72P Polymorphism in the Breast Cancer Patients of Ethnic Kashmiri Population Syeed N. World J Oncol [Internet] 2010 [cité 2022 mai 20];Available from: http://www.wjon.org/index.php/wjon/article/view/261..
- TP53 Pro47Ser and Arg72Pro polymorphisms and colorectal cancer predisposition in an ethnic Kashmiri population Sameer AS , Shah ZA , Syeed N , Banday MZ , Bashir SM , Bhat BA , Siddiqi MA . Genetics and molecular research: GMR.2010;9(2). CrossRef
- Evidence against involvement of p53 polymorphism in breast cancer predisposition Suspitsin EN , Buslov KG , Grigoriev MY , Ishutkina JG , Ulibina JM , Gorodinskaya VM , Pozharisski KM , et al . International Journal of Cancer.2003;103(3). CrossRef
- Intron 3 16 bp duplication polymorphism of p53 is associated with an increased risk for breast cancer by the age of 50 years Wang-Gohrke S, Becher H, Kreienberg R, Runnebaum IB , Chang-Claude J. Pharmacogenetics.2002;12(3). CrossRef
- TP53 PIN3 polymorphism associated with breast cancer risk in Iranian women Faghani M, Ghasemi FM , Nikhbakht M, Salehi M. Indian Journal of Cancer.2011;48(3). CrossRef
- CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies American Journal of Human Genetics.2004;74(6). CrossRef
- Linkage disequilibrium mapping of CHEK2: common variation and breast cancer risk Einarsdóttir K, Humphreys K, Bonnard C, Palmgren J, Iles MM , Sjölander A, Li Y, et al . PLoS medicine.2006;3(6). CrossRef
- Absence of CHEK2 1100delC, R145W and I157T Mutations in Breast Cancer in a Moroccan Population ElAmrani A, Moumad K, Attaleb M, Benhassou M, Försti A, Ennaji MM , Mzibri ME , Khyatti M. Journal of Cancer Research and Treatment.2014;2(1). CrossRef
- PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS Southey MC , Goldgar DE , Winqvist R, Pylkäs K, Couch F, Tischkowitz M, Foulkes WD , et al . Journal of Medical Genetics.2016;53(12). CrossRef
- The CHEK2 gene and inherited breast cancer susceptibility Nevanlinna H, Bartek J. Oncogene.2006;25(43). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2023
Author Details