Long non-coding RNA Panel on Prognosis of Glioma in Iranian Patients
Download
Abstract
Objective: Glioma is one of the most malignant brain tumors, accounting for about half of the gliomas that occur in central nervous system (CNS), originates from the glial tissue of the brain. The aim of the present study was to determine the expression levels of 3 lncRNAs (AGAP2-AS1, LINC01446 and HOTAIRM1) in patients with high grade glioma in comparison with low grade glioma.
Methods: Case group consisted of 70 high grade glioma patients control group consisted of 70 patients affected with low grade glioma. RNA extraction was performed using a RNA extraction kit.
Result: Our results showed that the expression of AGAP2-AS1 and LINC01446 genes significantly increased with increasing tumor grade (with fold-change ratio of 2.1 and 3.8 respectively). Also the expression of HOTAIRM1 gene increased with increasing tumor grade but this increase was not statistically significant (P value=0.6).
Conclusion: We concluded that AGAP2-AS1 and LINC01446 are promising lncRNA markers in prognosis of glioma.
Introduction
Glioma is a type of tumor that originates from the glial cells of the brain or the spine. Gliomas comprise about 30% of all brain and central nervous system tumors, and 80% of all malignant brain tumors. High-grade gliomas show extensive areas of necrosis and hypoxia and often, tumor growth causes a breakdown of the blood–brain barrier. As a rule, high-grade gliomas almost always grow back even after complete surgical excision, so are commonly called recurrent cancer of the brain [1-3]. Conversely, low-grade gliomas grow slowly, often over many years, and can be followed without treatment unless they grow and cause symptoms [4]. Glioma often occurs in the fourth to sixth decade of life, although some types are more common in children. Brain tumors are slightly more likely to occur in men. History of exposure to radiation is a risk factor for malignant glioma. Certain genetic disorders also increase the risk of developing these tumors in children, and rarely in adults. Several lifestyle risk factors have been investigated in relation to malignant glioma, including smoking or using mobile phones. The symptoms, prognosis and treatment of malignant glioma depend on the age of the diagnosis, the exact type of tumor and the location of the tumor in the brain. These tumors grow and invade normal brain tissue, which makes surgical removal very difficult, or sometimes impossible, and complicates treatment [5-7]. Several acquired genetic mutations have been found in gliomas. Tumor suppressor protein 53 (p53) is mutated early in the disease. p53 is the “guardian of the genome”, which, during DNA and cell duplication, makes sure the DNA is copied correctly and destroys the cell (apoptosis) if the DNA is mutated and cannot be fixed. When p53 itself is mutated, other mutations can survive. Phosphatase and tensin homolog (PTEN), another tumor suppressor gene, is itself lost or mutated. Epidermal growth factor receptor, a growth factor that normally stimulates cells to divide, is amplified and stimulates cells to divide too much. Together, these mutations lead to cells dividing uncontrollably, a hallmark of cancer. In 2009, mutations in IDH1 and IDH2 were found to be part of the mechanism and associated with a less favorable prognosis [8-10].
One of the factors that can help to increase the effectiveness of treatment is to better understand the molecular pathogenesis and genetic basis of gliomas. Previous studies have shown that long non-coding RNAs (lncRNA) are dysregulated in many malignant human tumors such as colorectal, prostate, bladder, hepatic, as well as cerebral. Recent evidence indicates that lncRNAs probably play an important role in the pathogenesis of gliomas. For example, these molecules can modulate cellular proliferation and apoptosis, leading to tumorigenesis. Studies also show that the abnormal expression of lncRNAs leads to a poor prognosis aggravates patients’ clinical condition, especially in GBMs. Therefore, lncRNAs seem to be applicable as potential diagnostic markers or therapeutic targets [11-13]. According to the aforementioned and the important role of lncRNAs (as potential diagnostic and therapeutic markers) in the pathogenesis of gliomas (regulating cellular proliferation and apoptosis, tumorigenesis) and their importance in determining GBM patients’ clinical outcomes and prognosis and risk of the disease, the aim of the present study was to investigate the expression profile of three lncRNAs including AGAP2-AS1, LINC01446 and HOTAIRM1 and their potential role as prognostic biomarkers in GBM.
Materials and Methods
In this study, 70 grade II or lower (as the control group) and 70 grade III or IV GBM tumors (as the case group) were collected from the patients who underwent surgery. All patients signed an informed consent form before surgery and agreed to their tissue samples being used in the research project. Tumor tissues were transferred into 1.5 mL sterile DNAse/RNAse free microtubes, and stored in -70 °C. RNA extraction was performed using a RNA extraction kit (MN Co, Germany, Cat No: 740304). Selected lncRNAs for the present study was AGAP2-AS1, LINC01446 and HOTAIRM1. Primer sequences used in this study to amplify target genes by Real-time PCR method showed in Table 1.
| Name of lncRNA | Primer sequence |
| AGAP2-AS1 | Forward: 5′‐TACCTTGACCTTGCTGCTCTC‐3′ |
| Reverse: 5′‐TGTCCCTTAATGACCCCAT CC‐3 | |
| LINC01446 | Forward: 5′‐AGAGCATACGGGAGAGATGAA-3′ |
| Reverse: 5′-AATTCTCCGAACGTGTCACGT-3′ | |
| HOTAIRM1 | Forward:5΄-GAAAGGCGAGCTTGGTTACGCTTAA-3′ |
| Reverse: 5΄-GACTTCGAAGCATTAACGATC-3΄ |
TaqMan probe real time PCR method used for specific amplification of lncRNAs and expression levels of these genes were compared using U6 as reference gene. PCR thermal program was as below: 95 °C for 5 minutes (Initial Denaturation), 95 °C for 30 seconds, 59 °C for 45 seconds, 72 °C for 30 seconds (repeat for 35 cycles) and 72 °C for 5 minutes (final extension).
Results
Demographic features of the studied population
Demographic information of assessed patients has been mentioned in Table 2.
| Variables | Mean ±SD (Range) | |
| Age (years) | 44.70 ±12.44 (42-77) (%) | |
| Grade | I | 5.3 |
| II | 31.9 | |
| III | 16.7 | |
| IV | 46.1 |
The mean age of patients was 44.70 ±12.44 years.
Gene expression findings
Changes in the expression of assessed genes in tumor tissues have been shown in Table 3.
| Name of lncRNA | Expression Change | Mean of fold change (ratio of Grade III&IV/Grade I&II) | P value |
| AGAP2-AS1 | Overexpressed | 2.1 | 0.001 |
| LINC01446 | Overexpressed | 3.8 | 0.001 |
| HOTAIRM1 | Overexpressed | 1.2 | 0.6 |
AGAP2-AS1 gene expression
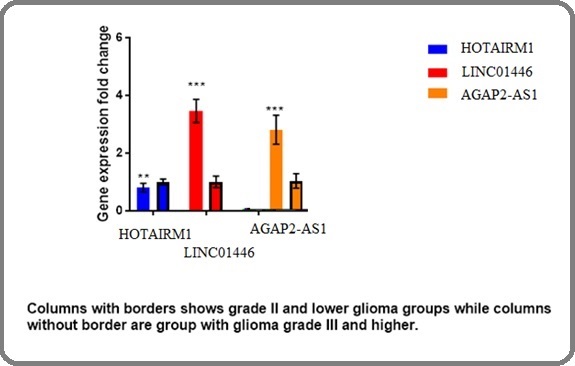
There was a statistically significant difference in AGAP2-AS1 gene expression between low- and high-grade tumors (p=0.001). The relative expression of this gene (fold change) was 2.1 in high grade (III and IV) respective to low grade (I and II) tumor tissues.
LINC01446 gene expression
Our findings showed that the expression of LINC01446 gene significantly increased in high grade tumors (p=0.001). The relative expression of this gene in grade III and IV samples was 3.8 folds higher compared to grade I and II tumors.
HOTAIRM1 gene expression
There was no association between the expression of HOTAIRM1 gene and tumor grade (p=0.45). The relative expression of this gene was 1.2-fold higher in high grade (III and IV) compared to low grade (I and II) samples (Figure 1).
Figure 1. The Relative Expression of Selected Genes in High- and Low-grade Gliomas.
Discussion
Recent microarray studies have revealed significant changes in the expression patterns of many lncRNAs in tissues of different subtypes of glioma and normal brain tissue. Using the lncRNA classification pipeline, Zhang et al. identified 1970 lncRNAs across different types and grades of human gliomas [14]. Wang et al. reported that maternally expressed gene 3 (MEG3) expression was markedly decreased in astrocytoma tissues compared to adjacent normal tissues. Recently, several studies have indicated that aberrant expression of lncRNAs may affect glioma initiation and progression [15]. Therefore, lncRNAs may act as biomarkers for glioma diagnosis, prognosis and target therapy [16]. AGAP2‐AS1, an antisense lncRNA, is located at human chromosome 12q14.1 and 1567 nucleotides in length [17]. Originally, AGAP2‐AS1 was identified as a novel lncRNA in lung cancer, and has found to be overexpressed in non‐small–cell lung cancer tissues [18]. Then, Fan et al also showed that the AGAP2‐AS1 expression was elevated in non‐small–cell lung cancer tissues, and served as a useful biomarker for discriminating non‐small–cell lung cancer tissues from normal lung tissues [18]. Ebrahimi et al, showed that the expression of ADAMTS9- AS2 and HOXA11-AS genes significantly increased with increasing tumor grade in Glioma patients. Also the expression of CASC2 gene significantly decreased with increasing tumor grade. They concluded that ADAMTS9-AS2 and HOXA11-AS and CASC2 are promising lncRNA markers in prognosis of glioma [19].
In conclusion, Our results showed that the expression of AGAP2-AS1 and LINC01446 genes significantly increased with increasing tumor grade (with fold-change ratio of 2.1 and 3.8 respectively). Also the expression of HOTAIRM1 gene increased with increasing tumor grade but this increase was not statistically significant (P value=0.6). We concluded that AGAP2-AS1 and LINC01446 are promising lncRNA markers in prognosis of glioma.
Ethics approval and consent to participate
All participants signed the informed consent form. The project was approved by the Local Research Ethics Committee (Shahid Beheshti medical university,Tehran, Iran).
Competing Interests
The authors declare that they have no competing interest.
Acknowledgements
The authors are grateful to all participants of the study. We are also thankful to Vice Chancellor for Research, Shahid Beheshti medical university, and Emam Hossein hospital (Tehran, Iran).
References
- Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma Lai NS , Wu DG , Fang XG , Lin YC , Chen SS , Li ZB , Xu SS . British Journal of Cancer.2015;112(7). CrossRef
- The 2007 WHO classification of tumours of the central nervous system Louis DN , Ohgaki H, Wiestler OD , Cavenee WK , Burger PC , Jouvet A, Scheithauer BW , Kleihues P. Acta Neuropathologica.2007;114(2). CrossRef
- The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: A systematic review Mansouri A, Mansouri S, Hachem LD , Klironomos G, Vogelbaum MA , Bernstein M, Zadeh G. Cancer.2016;122(16). CrossRef
- Survival in glioblastoma: a review on the impact of treatment modalities Delgado-López PD , Corrales-García EM . Clinical & Translational Oncology: Official Publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico.2016;18(11). CrossRef
- Long non-coding RNA in glioma: signaling pathways Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y, Wang S. Oncotarget.2017;8(16). CrossRef
- A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool Manterola L, Guruceaga E, Gállego Pérez-Larraya J, González-Huarriz M, Jauregui P, Tejada S, Diez-Valle R, et al . Neuro-Oncology.2014;16(4). CrossRef
- Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme Tan SK , Pastori C, Penas C, Komotar RJ , Ivan ME , Wahlestedt C, Ayad NG . Molecular Cancer.2018;17(1). CrossRef
- Accurate Delineation of Glioma Infiltration by Advanced PET/MR Neuro-Imaging (FRONTIER Study): A Diagnostic Study Protocol Verburg N, Pouwels PJW , Boellaard R, Barkhof F, Hoekstra OS , Reijneveld JC , Vandertop WP , Wesseling P, Witt Hamer PC . Neurosurgery.2016;79(4). CrossRef
- Hallmarks of cancer: the next generation Hanahan D, Weinberg RA . Cell.2011;144(5). CrossRef
- Tumor initiating cells in malignant gliomas: biology and implications for therapy Hadjipanayis CG , Van Meir EG . Journal of Molecular Medicine (Berlin, Germany).2009;87(4). CrossRef
- The emergent role of exosomes in glioma Gourlay J, Morokoff AP , Luwor RB , Zhu HJ , Kaye AH , Stylli SS . Journal of Clinical Neuroscience: Official Journal of the Neurosurgical Society of Australasia.2017;35. CrossRef
- miRNA Multiplayers in glioma. From bench to bedside Rolle K. Acta Biochimica Polonica.2015;62(3). CrossRef
- The functional role of long non-coding RNA in human carcinomas Gibb EA , Brown CJ , Lam WL . Molecular Cancer.2011;10. CrossRef
- Long non-coding RNA expression profiles predict clinical phenotypes in glioma Zhang X, Sun S, Pu JKS , Tsang ACO , Lee D, Man VOY , Lui WN , Wong STS , Leung GKK . Neurobiology of Disease.2012;48(1). CrossRef
- Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation Wang P, Ren Z, Sun P. Journal of Cellular Biochemistry.2012;113(6). CrossRef
- Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Cell Death & Disease.2016;7(5). CrossRef
- Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer Fan KJ , Liu Y, Yang B, Tian XD , Li CR , Wang B. European Review for Medical and Pharmacological Sciences.2017;21(10).
- Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer Qi F, Liu X, Wu H, Yu X, Wei C, Huang X, Ji G, Nie F, Wang K. Journal of Hematology & Oncology.2017;10(1). CrossRef
- Long non-coding RNA panel as a molecular biomarker in glioma Ebrahimi AA , Ashoori H, Vahidian F, Mosleh IS , Kamian S. Journal of the Egyptian National Cancer Institute.2021;33(1). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2023
Author Details