Decursin Inhibits Expression and Nuclear Localization of DNMT and HDAC, and Suppresses the Growth and Survival of Prostate Cancer Cells
Download
Abstract
Background: DNA methyltransferases (DNMT) and histone deacetylases (HDAC) inhibitors have utility in cancer therapeutics. However, they are associated with toxicity, which limits their efficacy against carcinomas. Previously, we reported that decursin has anticancer activity against prostate cancer (PCa). In this study, we investigated its epigenetic role in inhibiting the survival of PCa cells.
Methods and Results: PCa cell viability was evaluated using MTT and trypan blue assays. The effect of decursin on global methylation and gene-specific methylation was assessed by dot blot assay and MS-PCR, respectively. Differential expression of DNMTs and HDACs was analyzed through semi-quantitative RT-PCR and western blotting. The activity of these enzymes was measured using commercial kits. The nuclear localization of DNMTs and HDAC3 was assessed via immunoblotting.
Results: Decursin decreased cell viability and global methylation in PCa cells. It diminished the expression of DNMT1, DNMT3A, DNMT3B, and pan-DNMT activities. Decursin also reduced nuclear levels and increased cytosolic levels of DNMT1 and DNMT3A. Additionally, it inhibited the expression and activity of HDACs, leading to increased total acetylation and H3K9/14 acetylation of histone H3. A reduction in nuclear localization of HDAC3 was observed in decursin-treated PCa cells. Overall, decursin reduced DNMT and HDAC expression and enhanced the nuclear export of DNMT1, DNMT3A, and HDAC3, leading to decreased activities of these enzymes.
Conclusion: This study demonstrates the potent anticancer effects of decursin in prostate cancer cells through modulation of epigenetic mechanisms. Decursin decreased both overall and nuclear levels of DNMTs and HDACs. This effect is likely due to decursin’s inhibitory action on these enzymes, as indicated by decreased global methylation and increased histone H3 acetylation. These findings suggest that decursin has multi-faceted epigenetic modulatory properties, making it a promising therapeutic agent for prostate cancer prevention.
Introduction
Among the various reversible epigenetic changes, DNA methylation and histone deacetylation are the important modifications that contribute to cancer development [1, 2]. DNA methyl transferases (DNMTs) are the enzymes responsible for DNA methylation in cells. Gene-specific DNA hypermethylation and global hypomethylation are the two major events associated with cancer. Gene-specific hypermethylation of tumor suppressor genes results in the decreased expression of these genes, thus paving the way forward in the process of tumorigenesis [1, 3].
Genome-wide DNA hypomethylation is the other side of the coin which also contributes towards oncogenesis. It leads to the activation of certain retrotransposons [4] and oncogenes [5] which remain suppressed by methylation in normal cells. This activation culminates into different kinds of mutations in the genome leading to the tumorous transformation of the cells. Furthermore, hypomethylation causes genome instability and loss of imprinting, both of which have been reported to be linked with tumorigenesis [6, 7].
Expression levels of DNMTs also get dysregulated in several cancers and their over-expression has been observed to be associated with oncogenesis and aberrant methylation patterns. DNMT1, 3A, and 3B show increased expression in prostate cancer as compared to normal tissue [8]. DNMT inhibitors (DNMTi) are believed to revert the effect of gene-specific hypermethylation, thus reactivating the silenced tumor suppressor genes, ultimately leading to the reprogramming of cancer cells causing growth arrest and cell death [9].
Histone deacetylases (HDACs) catalyze the deacetylation of histone as well as non-histone proteins. HDACs have been reported to repress the level of an anti-proliferative gene, p21, and a pro-apoptotic gene, Bax [2]. HDACs also play a significant role in angiogenesis, invasion, migration, and differentiation by regulating various genes and transcription factors involved in these processes [2]. These chromatin regulators mediate their effect by deacetylation of histone proteins at the promoters of target genes, which leads to their transcriptional repression [2]. In clinical investigations, upregulation of HDACs is correlated with higher grade, increased proliferation rate, poor prognosis, and lower disease-free survival in most cases of cancer including prostate cancer [10, 11]. Thus, HDACs are being targeted for cancer therapeutics and HDAC inhibitors (HDACi) are being explored for their cancer therapeutic potential [12]. Combination therapies using both, DNMTi and HDACi have also been reported to be successful to some extent in certain cancers under experimental conditions and clinical trials [9]. Although, DNMTi and HDACi are still undergoing clinical trials, their higher cost, undesired toxicity after prolonged use, less bioavailability, and lower potency for solid tumors have limited their use in cancer therapeutics [12, 13]. Thus, the agents with dual effects as DNMTi and HDACi without causing any toxicity to the normal cells could be preferred for epigenetic therapy [14, 15]. Decursin, a coumarin compound (IUPAC name: [(3S)-2,2-dimethyl-8-oxo-3,4-dihydropyrano [3,2-g] chromen-3-yl] 3-methylbut-2-enoate and CID: 442126 ) isolated from Angelica gigas Nakai (AGN), has displayed anti-cancer properties against several cancers including prostate cancer, and found to be nontoxic to human prostate epithelial cell PWR-1E cells [16-21].
Decursin exhibits a diverse array of anti-cancer properties, including inhibition of cancer cell proliferation, induction of apoptosis, stimulation of autophagy, modulation of the immune system, and inhibition of invasion, metastasis, and angiogenesis. Its anti-proliferative effects are mediated by the induction of cell cycle arrest at various phases (G0/G1, S, and G2), by modulating upstream regulators such as p21, cyclin D1, and MAPK [18]. Decursin induces apoptosis by regulating mitochondrial membrane potential and various signaling molecules such as circle oxymase-2 (COX-2), caspases, survivin, Bax and Bcl-2 [22]. Decursin also modulates the expression of EMT associated proteins including N-Cadherin, E-Cadherin, and Vimentin, along with the pathways such as PI3K/AKT, ERK/JNK and p38 MAPK, to impede cancer cell migration. The targeted regulatory pathways can vary in different types of cells [22]. Additionally, it targets matrix metalloproteases to inhibit cancer cell invasion [20, 23] and downregulates VEGF to inhibit angiogenesis by altering ERK/JNK signaling [16, 22]. Moreover, decursin induces autophagy in various cancer cells [24] and enhances immune responses against cancer [22].
Specifically in prostate cancer cells, decursin induces growth inhibition, apoptosis, and cell cycle arrest at the G1 stage in DU145 and LNCaP cells, and at G1, S, and G2 stages in PC3 cells, with varied concentrations and treatment durations. Mechanistically, decursin upregulates p21 expression while downregulating CDK2/4/6 and cyclin D1 levels, consequently reducing CDK activity. Additionally, increased expression of cleaved caspase 3, caspase 9, and PARP elucidates the apoptotic mechanism induced by decursin [21].
However, there are no reports on whether decursin caused anticancer activities are mediated through modulation of DNMT and HDAC, which was investigated in the present study.
Materials and Methods
Cell culture and treatment
Human prostate cancer cells, LNCaP, DU145, and PC3 were from American Type Culture Collection (ATCC), USA, and cell repository at National Centre for Cell Science (NCCS), Pune, India. Cells were grown in RPMI 1640 media supplemented with 10% FBS and 1% of the antibiotic solution containing penicillin, streptomycin, and amphotericin B at standard conditions of 37ºC temperature, 95% humidified air, and 5% CO2. Cells were treated with DMSO control and desired concentrations of decursin (10-50 µM) for 72 h.
Cell proliferation and viability assays
MTT and Trypan blue assays were used for cell viability and proliferation analyses. For the MTT assay, cells were seeded at a density of 5,000 cells per well of a 96-well plate and after 72 h treatment with decursin (10, 25, and 50 µM), cell viability was analyzed using MTT as published earlier [25]. For Trypan blue assay 50,000 cells were seeded per well of a 6-well plate, treated accordingly, and counted after staining with trypan blue dye as described previously [25].
Global methylation analysis
Dot blot analysis was done to analyze the global methylation pattern by using an antibody against 5-methyl cytosine. DNA was isolated from the cells by phenol-chloroform extraction method and subjected to dot blot analysis. Fifteen nanograms of DNA were spotted on the nitrocellulose membrane. The DNA was cross-linked to the membrane by exposing it to ultraviolet radiation having energy of 1500 µJ/cm2 for 2-3 min. The membrane was then blocked in 5% BSA solution for 2 h followed by incubation with 5-methyl cytosine antibody at 4ºC overnight and then with HRP-conjugated secondary antibody for 2 h. The signal was detected by using ECL. The membrane was then counterstained with 0.2% methylene blue made in 0.5 M sodium acetate (pH 5) for 2 h.
Methylation-specific PCR
Methylation-specific PCR (MS-PCR) was performed to assess the promoter-specific methylation of certain genes. DNA from all the treatment groups was treated with bisulfite either manually [26] or by using an EZ DNA methylation lightning kit from Zymo Research (Cat. no. D5030) and then subjected to MS-PCR using gene-specific primers (Supplementary Table no.1).
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR)
RNA was isolated from the cells using Trizol solution followed by cDNA preparation and RT-PCR as per the standard protocol [25]. The bands were quantified by using Image J software and normalized against GAPDH. Primers for different genes are enlisted in Supplementary Table no. 2. Temperature profile used for RT-PCR was as follows: 95˚C for 5 min, 94˚C for 30 sec, X˚C for 30 sec, 72˚C for 1.30 min and 72˚C for 5 min. Steps 2-4 were operated for 25-30 cycles. X indicates the annealing temperature.
Western blot analysis
Cells were treated with decursin (10, 25, and 50 µM) for 72 h followed by preparation of whole cell lysate and nuclear/cytosolic fraction followed by western blot analysis as reported earlier [27]. Bands were quantified by Image J software and normalized to loading control.
DNMT activity assay
The activity of DNMT was assessed by using an EpiQuik DNMT activity/inhibition assay kit (Cat no. P-3001-1) as per the protocol mentioned in the manual. Fifteen µg of nuclear lysate was taken as input material.
HDAC activity assay
The activity of HDAC was measured by an EpiQuik HDAC activity/inhibition assay kit (cat no. P-4002-48) following the manufacturer’s protocol. Twelve µg of nuclear lysate was used as input material.
Statistical analysis
Statistically, the significant difference between control and drug treatment groups was assessed by Student’s t-test using GraphPad Prism version 5. The p-value ≤ 0.05 was considered significant. Image J software was used to quantify the western blots and semi-quantitative RT-PCR gel images.
Results
Decursin inhibits the growth and viability of prostate cancer cells in combination with DNMT and HDAC inhibitors
Prostate carcinoma DU145 cells were treated with decursin (10-50 µM) alone and in combination with DNMT inhibitor, 5-aza, 2-deoxycytidine (DAC, 15 µM), or HDAC inhibitor, TSA (0.2 µM) for 72 h followed by MTT and trypan blue dye exclusion assays (Figure 1).
Figure 1. Effect of Decursin alone and in Combination with DNMT Inhibitor, DAC, and HDAC Inhibitor, TSA on Cell Viability of DU145 Cells. (A) Chemical structure of decursin showing the coumarin ring. (B) and (C) Cell viability analysis by MTT and trypan blue exclusion assay, respectively. DU145 cells were treated with DMSO vehicle control or 10, 25, and 50 µM concentration of decursin or 0.2 µM concentration of TSA or a combination of decursin (25 µM) and TSA (0.2 µM) for 72 h followed by MTT (B) as well as trypan blue (C) assays were performed for cell viability analysis. The data are presented as mean ± SEM of three independent experiments. * P<0.05, **P<0.01, ***P<0.001 versus control. DAC; 5-aza-deoxycytidine, DN, decursin; TSA, trichostatin A.
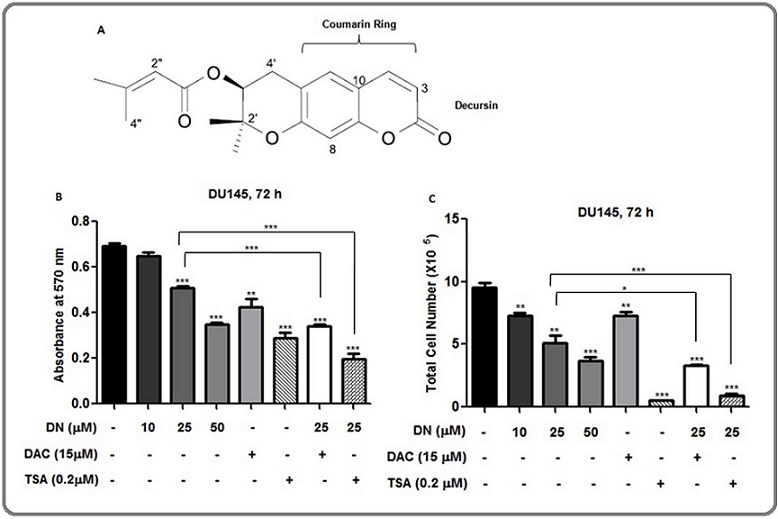
Consistent with our previous report, decursin (Figure 1A) decreased the cell viability in a concentration-dependent manner [21]. The enhanced growth inhibition (P < 0.05-0.001) was observed in combination treatment of decursin (25 µM) and DAC (Figure 1B and 1C). The 50 µM concentration of decursin showed comparable decrease in cell viability to that of TSA. Similarly, a strong increase in growth inhibition (P<0.001) was observed with the combination treatment of decursin (25 µM) and TSA (Figure 1B and 1C). Next, we investigated whether decursin could modulate the expression and/or activity of epigenetic enzymes, DNMTs, and HDACs.
Effect of decursin on global methylation in prostate cancer cells
A dot blot assay was performed to analyze the effect of decursin on global methylation. Higher concentrations of decursin showed decrease in the levels of global methylation in DU145 and LNCaP cells. A respective decrease of 41% and 46% was observed at 25 µM and 50 µM concentrations of decursin in DU145 cells while this decrease was 29% and 37% in LNCaP cells, respectively (Figure 2A).
Figure 2. Effect of Decursin on Global Methylation and Gene-specific Methylation of Certain Genes in Prostate Cancer Cells. (A) Dot blot analysis, (B-D) Methylation-specific PCR. Cells were treated with DMSO vehicle control or 10, 25, and 50 µM concentrations of decursin or 5 μM concentration of DAC for 96 h, and after the completion of treatment time, DNA was isolated from cells and dot blot analysis was done by using 5-methyl cytosine antibody. The membrane was then counterstained with methylene blue as loading control. The blots were quantified by image J and normalized against loading control. Blot intensity is mentioned below each blot as fold change from control (A). For gene-specific methylation assessment, cells were treated with DMSO vehicle control or 50 µM concentration of decursin for 96 h. DNA was isolated from the cells and subjected to methylation-specific PCR and PCR products were run on 3% agarose gel (B-D). The band intensities of unmethylated and methylated bands were quantified by image J and represented as U/M ratio graphs below the gel images (Table A-C). ns, non-significant, **P0<.01, ****P<0.0001 versus control. DN, decursin; DAC, 5-aza, 2-deoxy cytidine.
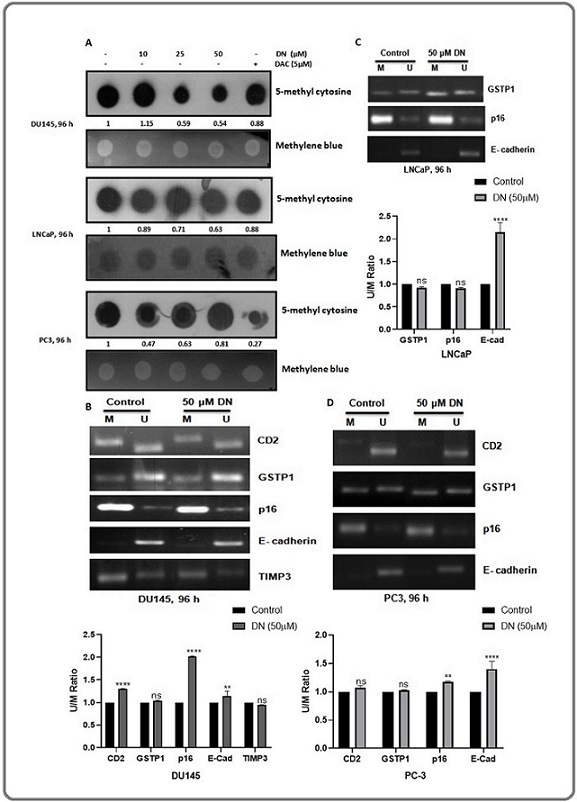
In PC3 cells, the highest decrease of 53% was observed at 10 µM concentration of decursin, whereas, 25 and 50 µM decursin decreased the global methylation by 37% and 19%, respectively (Figure 2A). Thus, all the three cell lines showed a decrease in the global methylation after decursin treatment, hence reflecting a possibility of DNMT inhibition by decursin.
Effect of decursin on promoter-specific methylation in prostate cancer cells
Methylation-specific PCR (MS-PCR) was utilized to study the effect of decursin on the promoter-specific methylation of certain genes which are usually found to be hypermethylated in different types of cancers including prostate cancer. A ratio of unmethylated/ methylated band intensity (U/M) was calculated for easier analysis and normalized with respect to control. The methylation level of the CD2, p16 and E-Cadherin gene was decreased remarkably in DU145 cells after the treatment with a 50 µM concentration of decursin as indicated by the increased U/M ratio while no significant change in the methylation status of GSTP1 and TIMP3 gene was observed in this cell line (Figure 2B). No noticeable effect was seen in the methylation status of GSTP1 and p16 gene in LNCaP cells, while the methylation level of E-cadherin was decreased significantly in this cell line (Figure 2C). In PC3 cells, methylation levelof p16 and E-Cadherin was decreased significantly after decursin treatment while no significant change was observed in the methylation status of CD2 and GSTP1 gene (Figure 2D). Overall, decursin showed a decrease in the methylation level of the CD2, p16 and E-Cadherin gene in DU145 cells, E-Cadherin gene in LNCaP cells and p16 & E-Cadherin gene in PC3 cells.
Decursin downregulates the expression of DNMTs in prostate cancer cells
As decursin treatment resulted in the decreased genome-wide methylation and promoter-specific methylation of p16, we studied the effect of decursin on the mRNA and protein expression of DNMTs. Both the cell lines, DU145 and LNCaP displayed a decrease in the mRNA expression levels of DNMTs at higher concentrations of decursin. There was a moderate decrease in the expression of DNMT1 while a prominent decrease was observed in the expression of DNMT3A and 3B in DU145 cells. The lowest concentration of decursin (10 µM) showed variable effects on the mRNA expression of DNMT1, and DNMT3A and 3B (Figure 3A). A noticeable decrease was observed in the expression of DNMT1 and DNMT3B by decursin in LNCaP cells (Figure 3B).
Figure 3. Effect of Decursin on mRNA and Protein Expression of DNMTs in Prostate Cancer Cells. (A, B) Representative agarose gel electrophoresis images of semi-quantitative RT-PCR showing differential mRNA expression of DNMT1, 3A, 3B in DU145 and LNCaP cells after treatment with different concentrations of decursin for 72 h. (C, D) Western blots signifying differential protein levels of DNMTs in DU145 and LNCaP cells after treatment with various concentrations of decursin for 72 h. DN, decursin; DNMT: DNA methyl transferase, GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
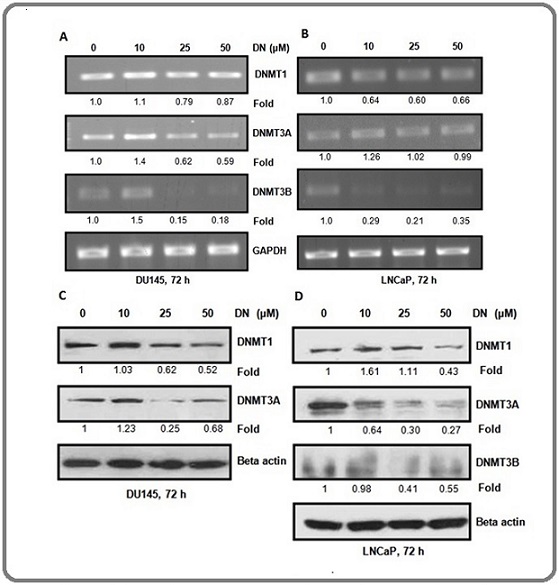
Further, protein level of DNMT1 and 3A was decreased after decursin treatment with the exception of l0 µM concentration in DU145 cells (Figure 3C). In LNCap cells, protein expression of DNMT1 was found to be decreased only at the highest concentration of decursin while the lower concentrations exhibited a slight increase. The protein expression of DNMT3A and DNMT3B was decreased after decursin treatment (Figure 3D). Overall, the results exhibited a decrease in mRNA and protein expression of DNMTs in both the prostate cancer cell lines after decursin treatment.
Decursin downregulates the expression of HDACs and increases the total acetylation of H3 and acetylation of H3 at lysine 9/14 in prostate cancer cells
Furthermore, to explore the possibility that the inhibition of the cancer cell viability by decursin was contributed by the diminished expression HDACs, we analyzed mRNA and protein expression levels of HDACs.
The mRNA level of HDAC1, HDAC2, and HDAC3 was decreased in a concentration-dependent manner in DU145 cells except for 10 µM concentration of decursin (Figure 4A). However, there was no change in the mRNA level of HDACs by decursin in LNCaP cells (Figure 4B).
Figure 4. Effect of Decursin on mRNA and Protein Expression of HDACs and Protein Expression of Acetylated H3K9/14 and Total Acetylation of H3 in DU145 and LNCaP Cells. (A,B) Representative agarose gel electrophoresis images of semi-quantitative RT-PCR showing differential mRNA expression of HDAC1, 2, and 3 in DU145 and LNCaP cells after treatment with different concentrations of decursin for 72 h. (C, D) Western blots signifying differential protein levels of HDACs in DU145 and LNCaP cells after treatment with various concentrations of decursin for 72 h. (E) Western blot bands for total acetylation (K9/14/18/23/27) of histone H3 after treatment with various concentrations of decursin for 72 h. (F,G) Western blots showing the effect of decursin on acetylation level of H3K9/14 in DU145 and LNCaP cells after treatment with various concentrations of decursin for 72 h DN. DN, decursin; HDAC, histone deacetylase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
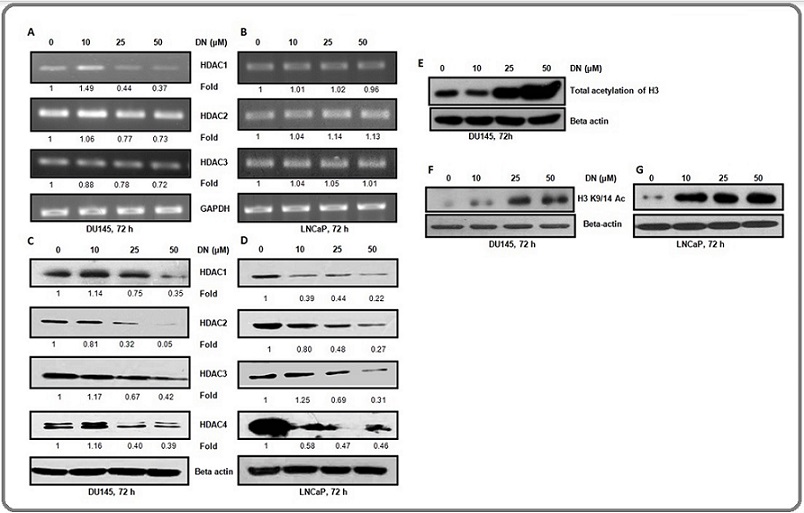
The protein levels of all HDACs diminished in a concentration-dependent manner in DU145 cells (Figure 4C). There was a concentration-dependent decrease in the protein levels of HDAC1, 2, 3, and 4 in LNCaP cells (Figure 4D). The results exhibited a significant decrease in protein expression of HDACs in both the cell lines after decursin treatment. Next, we analyzed the effect of decursin on the protein level of total acetylation (K9, K14, K18, K23, and K27) of histone 3 by western blotting and found that the total acetylation level of H3 increased after decursin treatment in DU145 (Figure 4E). Furthermore, we checked the specific acetylation of histone H3 at lysine 9/14(H3K9/14 Ac) and it was also increased after decursin treatment in both the cell lines (Figure 4F and G). These findings corroborated the observed decrease in the protein expression level of HDACs after decursin treatment.
Decursin reduced the overall DNMT and HDAC activity in DU145 and LNCaP cells
The effect of decursin treatment on pan-DNMT and pan-HDAC enzymatic activity was analyzed in DU145 and LNCaP cells and the overall activity of DNMTs was decreased in a concentration-dependent manner in both the cell lines. At the highest concentration of decursin, the inhibitory activity was found to be comparable to that of a known inhibitor, DAC indicating a great potential of decursin as an inhibitor of DNMT activity (Figure 5A and B).
Figure 5. Inhibition of pan-DNMT and pan-HDAC Activity by Decursin in Prostate Cancer Cells. (A,B) Graphical representation of the percentage of DNMT activity in DU145 and LNCaP cells, respectively. (C,D) Graphical representation of HDAC activity in DU145 and LNCaP cells. Nuclear lysate of cells was prepared after treatment of cells with DMSO control or different concentrations of decursin for 72 h and subjected to activity analysis by EpiQuik DNMT Activity/Inhibition Assay kit and EpiQuik HDAC Activity/Inhibition Assay Kit, respectively. The data are presented as mean ± SEM of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 versus control. DAC: 5-Aza, deoxycytidine, DN, decursin; DNMT: DNA methyl transferase, HDAC, histone deacetylase.
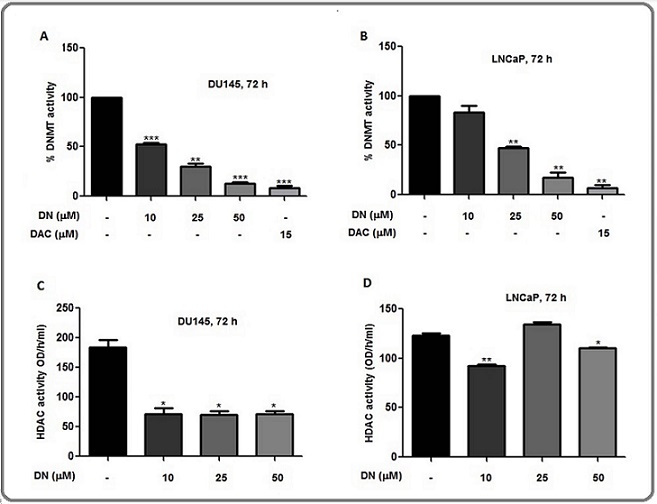
Furthermore, the pan-HDAC activity was significantly decreased at all the concentrations (10, 25, and 50 µM) of decursin in DU145 cells (Figure 5C). In LNCaP cells, activity was decreased at 10 µM and 50 µM concentrations while no significant effect was observed at 25 µM concentration and the highest effect was observed at 10 µM concentration (Figure 5D). As a result, decursin not only decreased the expression levels of DNMTs and HDACs but also diminished their activity in both cell lines.
Decursin decreases the nuclear localization of DNMTs and HDACs
The effect of decursin on the nuclear localization of DNMTs and HDACs was evaluated as a part of examining the possible mechanisms of decursin-mediated reduction in their activities. A concentration-dependent decrease was observed in the nuclear levels of all the three DNMTs in DU145 cells while the cytoplasmic levels of DNMT1 and DMNT3A were found to be increased at all the concentrations of decursin as compared to control. Nuclear levels of DNMT3B were decreased at all the used concentrations of decursin (Figure 6A).
Figure 6. Nuclear Levels of DNMTs and HDAC3 after Decursin Treatment in Prostate Cancer Cells. (A-C). Representative blots showing nuclear localization of DNMTs in DU145 cells, and HDAC3 in DU145 and LNCaP cells, respectively after treatment with different concentrations of decursin (10, 25, and 50 µM) for 72 h. DN, decursin; HDAC, histone deacetylase; DAPI, 4', 6-diamidino-2-phenylindole. Histone H3 and beta actin are used as nuclear and cytosolic loading controls, respectively.
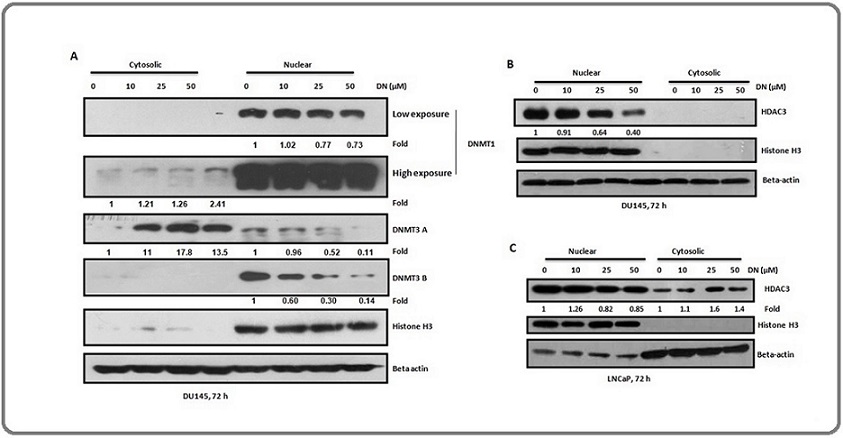
These observations suggest that decursin decreased the nuclear localization of DNMTs and may export these enzymes out of the nucleus, thus decreasing their activity inside the nucleus.
Further, western blotting for HDAC3 using the nuclear and cytosolic lysates of DU145 cells showed a decrease in the nuclear level of HDAC3 after decursin treatment in a concentration-based manner (Figure 6B). Similarly, in LNCaP cells, the nuclear level of HDAC3 was decreased after decursin treatment with a corresponding increase in its cytosolic level (Figure 6C). Overall, results indicated that decursin has the ability to decrease the nuclear localization of DNMTs and HDACs which may in part be associated with its ability to inhibit the activities of these enzymes.
Discussion
The central findings in the present study using human prostate cancer cells are that decursin (a) inhibited global methylation and promoter methylation of tumor suppressor genes, (b) suppressed the mRNA and protein expression of DNMT1, DNMT3A, and DNMT3B, (c) decreased the nuclear localization and activities of DNMTs, (d) down-regulated the expression of HDACs as well as their nuclear localization, (e) increased the total acetylation of histone H3 and acetylation of H3K9/14 (f) decreased the growth and survival of prostate cancer cells which were further enhanced in combination with DNMTs and HDACs inhibitors.
Epigenetic enzymes have become vital targets in cancer therapeutics; however, the use of clinically approved inhibitors against these enzymes is limited due to their shortcomings including non-specific toxicity and application at the early stages [10]. Thus, novel agents are being explored which can overcome these shortcomings and provide therapeutic benefits with greater efficacy. In such cases, small molecules like decursin may have some advantage as it targets multiple pathways at genetic as well as epigenetic levels and is also non-toxic to normal cells. Decursin has been earlier tested for its toxicity in normal prostate epithelial cells and has been found to be non-toxic in these cells [21], and decursin is also reported to have anticancer properties in prostate cancer cells and animal models [28]. Nevertheless, the mechanisms behind the anti-cancer activity of decursin against prostate cancer are yet to be deciphered completely. Our present study suggested the novel DNMT and HDAC inhibitory potentials of decursin in prostate cancer cells.
Decursin manifested a decrease in global methylation in all the three prostate cancer cell lines. As global hypomethylation may also lead to genomic instability and cell death [6], the decursin-mediated decrease in cancer cell viability may in part be due to genomic instability resulted due to the decreased global methylation by decursin. For gene-specific methylation analysis, certain genes which are commonly methylated in prostate cancer cells were studied. The p16 gene is an important tumor suppressor gene that is commonly found methylated and down-regulated in prostate cancer [29]. Thus, the observation of decursin-mediated hypomethylation of the p16 gene in DU145 and PC3 cells could be one of the factors contributing to the decrease in cell growth and viability of cancer cells by decursin. This could be supported by the report in which the inhibition of methylation at the p16 gene promoter caused growth suppression in various cancer cells [30]. Cyclin D2 either acts as a tumor suppressor gene or oncogene depending upon the cancer subtype. In prostate cancer, its silencing by hypermethylation is associated with tumor aggressiveness and a high Gleason score [31]. Previous studies showed that treatment of prostate cancer cells with DAC and TSA increased the Cyclin D2 expression which was accompanied by the decreased proliferation of LNCaP cells [32]. Our results exhibited a significant decrease in the methylation level of Cyclin D2 promoter in DU145 cells after decursin treatment which may also contribute to the decursin-mediated anti-cancer effects. E-Cadherin regulates cell adhesion and migration and thus, it inhibits epithelial to mesenchymal transition (EMT) of cancer cells reflecting its tumor suppressorive nature. It is reported to get silenced in various types of cancers by promoter hypermethylation [33] including prostate cancer to enhance the invasiveness of cancer cells [34]. Khan et al reported that a treatment with a phytochemical, EGCG resulted in the decreased levels of methylation of E-Cadherin in cervical cancer cells leading to the restoration of its expression [33]. In our study, we also observed the decreased methylation levels of E-Cadherin promoter in all the three prostate cancer cell lines after decursin treatment.
Furthermore, we found that decursin downregulated the mRNA as well as the protein expression of DNMTs and HDACs in cancer cells. However, only protein levels of DNMT3A and all HDACs were found altered in LNCaP cells. Since DU145 cells are androgen- independent and LNCaP cells are androgen-dependent, therefore, the possibility of differential regulation of DNMTs and HDACs could be associated with their advanced and androgen-independent state versus less aggressive and androgen-dependent stage of the cancer cells. Small molecules such as 3,3’-diindolylmethane (DIM) and parthenolide have been reported to decrease the protein expression of HDACs by proteasomal degradation [35]. The decreased protein expression of HDAC3 after sulforaphane treatment was reported due to pin1-mediated proteasomal degradation of HDAC3 [36]. Similarly, in addition to transcriptional and translational regulations, the decursin-mediated decrease in the protein expression of DNMTs and HDACs may involve such post-transcriptional mechanisms which require further investigation.
In the present study, we observed a decrease in the expression as well as activity of DNMTs after decursin treatment. Similarly, phytochemicals such as rosmarinic acid, apigenin, sulforaphane, EGCG, genistein, and curcumin, etc have been shown to decrease the expression as well as activity of DNMTs [37]. DNMTs regulate gene expression by methylating DNA, and thus, their activity is mostly restricted to the nucleus. Apart from this, the decreased levels of nuclear DNMTs are associated with their mislocalization in cytoplasm. For instance, DNMT1 mostly resides in the nucleus with an exception during preimplantation period of embryonic development. In this stage, DNMT localizes in the cytoplasm. Apart from this stage, cytoplasmic localization of DNMT1 is associated with various diseases [38]. A long non coding RNA,CCD26 has been found to interact with DNMT1 and ensure its nuclear localization. Knockout of this RNA is reported to cause cytoplasmic localization of DNMT, possibly through nucleus to cytoplasmic export of DNMT1 [38]. Consistent with this, In our study, decursin was found to reduce the nuclear localization of DNMT1 and DNMT3A with a corresponding increase in their cytoplasmic levels. This result indicated that the decreased nuclear localization of DNMTs by decursin could contribute to the inhibitory effect of decursin on DNMT activity in prostate cancer cells.
Since, decursin displayed HDAC inhibitory effect in prostate cancer cells, we further analyzed its effect on histone acetylation. The increase in the total acetylation level of histone H3 and acetylation of H3K9/14 after decursin treatment further validated its effect as HDAC inhibitor. This was further supported by measuring the pan-enzymatic activity of HDACs after decursin treatment through a colorimetric activity assay. As anticipated, decursin reduced the overall activity of HDACs in both the prostate cancer cell lines. Other previously reported small molecules such as BITC and sulforaphane also displayed similar effects on the protein expression as well as the activity of HDACs [35]. Further, our findings also suggested the decreased nuclear localization of HDAC3 by decursin treatment. Thus, this data concluded that decursin may act as a potential inhibitor of HDAC activity.
Despite having non-histone targets, the role of HDACs in cancer development is mainly associated with the deacetylation of histones at the promoter of various genes such as p21, Bax, and E-cadherin, etc in the nucleus [2]. HDAC inhibitors which are being explored as anti-cancerous agents induce histone H3 and H4 acetylation at the promoters of these genes [2, 39, 40].
It suggests that the nuclear localization of HDACs has a role in carcinogenesis, and a chemical showing HDAC inhibitory activity may also decrease the nuclear level of HDACs. Hence, the decursin-caused reduced nuclear localization of HDAC3 suggested its role in modulating the cellular versus nuclear localization of HDACs. Consequently, the reduction in the nuclear localization of HDACs could be one of the possible mechanisms by which decursin inhibited the activity of HDAC. This speculation is supported by a study carried out by Rajendran et al. where Sulforaphane treatment of colon cancer was reported to cause increased phosphorylation of HDAC3 and its interacting partner SMRT, thus causing dissociation of this complex. This was further resulted in binding of HDAC3 by 14-3-3 and pin 1, ultimately leading to the trafficking of HDAC3 from nucleus to cytoplasm and this model was the proposed mechanism behind the inhibitory role of sulforaphane in colon cancer cells [36]. Decursin also decreased the mRNA levels of other epigenetic enzymes such as MLL1, EZH2, LSD, and p300 (Supplementary Figure 1), hence, may emerge an important epigenetic modulator for its anti-tumor effects. In conclusion, the ability of decursin to decrease the overall expression and nuclear levels of DNMTs and HDACs may contribute to its DNMT and HDAC inhibitory roles in prostate cancer as reflected by the decreased activity of these enzymes (Figure 7).
Figure 7. Schematic Representation of Epigenetic Modifications Induced by Decursin in Prostate Cancer Cells. Decursin decreased the overall expression and activity of two major epigenetic modifiers DNMTs and HDACs in DU145 and LNCaP cells. It further decreased the nuclear levels of DNMTs and corresponding increase in the cytosol was observed in DU145 cells. Similarly, decursin mediated decrease in nuclear levels of HDACs and corresponding increase in cytosol was observed in LNCaP cells. Further, as a result of decreased expression of HDACs in nucleus, elevated acetylation levels of histone H3 were found in both the cell lines. Image was created using biorender software.
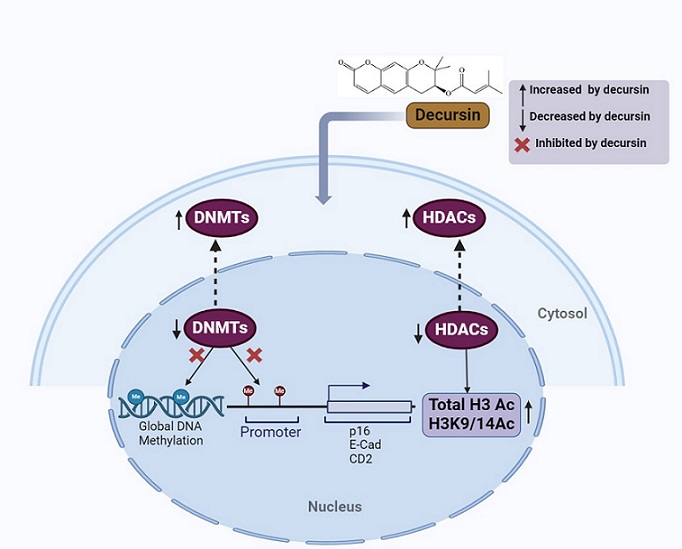
Conclusively, the study finds decursin as a novel, non-toxic epigenetic modulator targeting both DNMTs and HDACs for inhibiting the growth and survival of prostate cancer cells.
Declarations
Consent For Publication
Not applicable
Acknowledgements
This work was supported financially by the Department of Science and Technology (DST)-PURSE, University Grants Commission (UGC)-RNW and UPE-II, JNU, DBT-Builder, DRS-UGC, India grants. Lalita is thankful to UGC, India for providing Junior and Senior Research Fellowships.
Authors Contribution
Conceptualization, R.P.S and P.R.; methodology, L.D, R.P.S, R.P, A.J and S.K.; formal analysis, L.D and R.P.; resources, R.P.S; writing - original draft preparation, L.D; writing - review and editing, L.D, A.J, D.Y. and R.P.S.; Providing decursin, D.Y.; project administration and funding acquisition, R.P.S. All authors have read and agreed to the published version of the manuscript.
Conflict Of Interest
The authors declare that no competing interests exist.
References
- DNA methylation and cancer Das PM , Singal R. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2004;22(22). CrossRef
- Histone deacetylases and cancer Glozak M. A., Seto E.. Oncogene.2007;26(37). CrossRef
- A decade of exploring the cancer epigenome - biological and translational implications Baylin SB , Jones PA . Nature Reviews. Cancer.2011;11(10). CrossRef
- Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features Cho N.-Y., Kim B.-H., Choi M., Yoo E. J., Moon K. C., Cho Y.-M., Kim D., Kang G. H.. The Journal of Pathology.2007;211(3). CrossRef
- Epigenetic changes in prostate cancer: implication for diagnosis and treatment Li L, Carroll PR , Dahiya R. Journal of the National Cancer Institute.2005;97(2). CrossRef
- Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK , Liloglou T. International Journal of Cancer.2009;124(1). CrossRef
- Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues Jarrard D. F., Bussemakers M. J., Bova G. S., Isaacs W. B.. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.1995;1(12).
- DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer Kobayashi Y, Absher DM , Gulzar ZG , Young SR , McKenney JK , Peehl DM , Brooks JD , Myers RM , Sherlock G. Genome Research.2011;21(7). CrossRef
- Recent developments in epigenetic cancer therapeutics: clinical advancement and emerging trends Nepali K, Liou J. Journal of Biomedical Science.2021;28(1). CrossRef
- Histone deacetylases and cancer Barneda-Zahonero B, Parra M. Molecular Oncology.2012;6(6). CrossRef
- Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy Weichert W., Röske A., Gekeler V., Beckers T., Stephan C., Jung K., et al . British Journal of Cancer.2008;98(3). CrossRef
- Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs Kim H, Bae S. American Journal of Translational Research.2011;3(2).
- Insights into the molecular interactions of thymoquinone with histone deacetylase: evaluation of the therapeutic intervention potential against breast cancer Parbin S, Shilpi A, Kar S, Pradhan N, Sengupta D, Deb M, Rath SK , Patra SK . Molecular bioSystems.2016;12(1). CrossRef
- Epigenetic targets in cancer and aging: dietary and therapeutic interventions Khan S, Shukla S, Sinha S, Meeran SM . Expert Opinion on Therapeutic Targets.2016;20(6). CrossRef
- Silibinin synergizes with histone deacetylase and DNA methyltransferase inhibitors in upregulating E-cadherin expression together with inhibition of migration and invasion of human non-small cell lung cancer cells Mateen S, Raina K, Agarwal C, Chan D, Agarwal R. The Journal of Pharmacology and Experimental Therapeutics.2013;345(2). CrossRef
- Decursin and decursinol angelate inhibit VEGF-induced angiogenesis via suppression of the VEGFR-2-signaling pathway Jung MH , Lee SH , Ahn E, Lee Y. Carcinogenesis.2009;30(4). CrossRef
- Decursin exerts anti-cancer activity in MDA-MB-231 breast cancer cells via inhibition of the Pin1 activity and enhancement of the Pin1/p53 association Kim J, Jung JH , Kim S, Jeong S. Phytotherapy research: PTR.2014;28(2). CrossRef
- Decursin inhibits growth of human bladder and colon cancer cells via apoptosis, G1-phase cell cycle arrest and extracellular signal-regulated kinase activation Kim W, Lee S, Choi Y, Moon S. International Journal of Molecular Medicine.2010;25(4). CrossRef
- Decursin and decursinol from Angelica gigas inhibit the lung metastasis of murine colon carcinoma Son SH , Park K, Park SK , Kim YC , Kim YS Y, Lee S, Chung W. Phytotherapy research: PTR.2011;25(7). CrossRef
- Decursin and decursinol inhibit VEGF-induced angiogenesis by blocking the activation of extracellular signal-regulated kinase and c-Jun N-terminal kinase Son SH , Kim M, Chung W, Son J, Kim Y, Kim Y, Kang SS , Lee S, Park K. Cancer Letters.2009;280(1). CrossRef
- A novel anticancer agent, decursin, induces G1 arrest and apoptosis in human prostate carcinoma cells Yim D, Singh RP , Agarwal C, Lee S, Chi H, Agarwal R. Cancer Research.2005;65(3).
- A comprehensive review of the anticancer effects of decursin Chu Y, Yuan Q, Jiang H, Wu L, Xie Y, Zhang X, Li L. Frontiers in Pharmacology.2024;15. CrossRef
- Decursin prevents TPA-induced invasion through suppression of PKCα/p38/NF-κB-dependent MMP-9 expression in MCF-7 human breast carcinoma cells Kim J, Noh E, Kim M, Hwang J, Hwang H, Ryu D, Kim H, et al . International Journal of Oncology.2014;44(5). CrossRef
- Decursin inhibits cell growth and autophagic flux in gastric cancer via suppression of cathepsin C Kim S, Lee S, Kim N, Joo M, Lee K, Lee M, Jeon HJ , et al . American Journal of Cancer Research.2021;11(4).
- Fisetin inhibits various attributes of angiogenesis in vitro and in vivo--implications for angioprevention Bhat TA , Nambiar D, Pal A, Agarwal R, Singh RP . Carcinogenesis.2012;33(2). CrossRef
- Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands Herman J. G., Graff J. R., Myöhänen S., Nelkin B. D., Baylin S. B.. Proceedings of the National Academy of Sciences of the United States of America.1996;93(18). CrossRef
- Silibinin inhibits aberrant lipid metabolism, proliferation and emergence of androgen-independence in prostate cancer cells via primarily targeting the sterol response element binding protein 1 Nambiar DK , Deep G, Singh RP , Agarwal C, Agarwal R. Oncotarget.2014;5(20). CrossRef
- Prostate Cancer Xenograft Inhibitory Activity and Pharmacokinetics of Decursinol, a Metabolite of Angelica gigas Pyranocoumarins, in Mouse Models Wu W, Tang S, Zhang Y, Puppala M, Cooper TK , Xing C, Jiang C, Lü J. The American Journal of Chinese Medicine.2017;45(8). CrossRef
- Association of p16 gene methylation with prostate cancer risk: a meta-analysis Feng W, Han Z, Zhu R, Liu P, Liu S. Journal of B.U.ON.: official journal of the Balkan Union of Oncology.2015;20(4).
- Inhibition of DNA methylation by 5-aza-2'-deoxycytidine suppresses the growth of human tumor cell lines Bender C. M., Pao M. M., Jones P. A.. Cancer Research.1998;58(1).
- Hypermethylation of Cyclin D2 is associated with loss of mRNA expression and tumor development in prostate cancer Henrique R, Costa VL , Cerveira N, Carvalho AL , Hoque MO , Ribeiro FR , Oliveira J, et al . Journal of Molecular Medicine (Berlin, Germany).2006;84(11). CrossRef
- Restoration of cyclin D2 has an inhibitory potential on the proliferation of LNCaP cells Kobayashi T, Nakamura E, Shimizu Y, Terada N, Maeno A, Kobori G, Kamba T, et al . Biochemical and Biophysical Research Communications.2009;387(1). CrossRef
- (-)-Epigallocatechin-3-gallate reverses the expression of various tumor-suppressor genes by inhibiting DNA methyltransferases and histone deacetylases in human cervical cancer cells Khan MA , Hussain Arif, Sundaram MK , Alalami U, Gunasekera D, Ramesh L, Hamza A, Quraishi U. Oncology Reports.2015;33(4). CrossRef
- DNA methylation of E-cadherin is a priming mechanism for prostate development Keil KP , Abler LL , Mehta V, Altmann HM , Laporta J, Plisch EH , Suresh M, Hernandez LL , Vezina CM . Developmental Biology.2014;387(2). CrossRef
- Dietary phytochemicals, HDAC inhibition, and DNA damage/repair defects in cancer cells Rajendran P, Ho E, Williams DE , Dashwood Rh . Clinical epigenetics.2011;3(1). CrossRef
- Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly Rajendran P, Delage B, Dashwood WM , Yu T, Wuth B, Williams DE , Ho E, Dashwood RH . Molecular Cancer.2011;10. CrossRef
- Dietary phytochemicals as epigenetic modifiers in cancer: Promise and challenges Shankar E, Kanwal R, Candamo M, Gupta S. Seminars in Cancer Biology.2016;40-41. CrossRef
- A long intergenic non-coding RNA regulates nuclear localization of DNA methyl transferase-1 Jones R, Wijesinghe S, Wilson C, Halsall J, Liloglou T, Kanhere A. iScience.2021;24(4). CrossRef
- Dietary agents as histone deacetylase inhibitors: sulforaphane and structurally related isothiocyanates Dashwood RH , Ho E. Nutrition Reviews.2008;66 Suppl 1(Suppl 1). CrossRef
- Green tea polyphenols causes cell cycle arrest and apoptosis in prostate cancer cells by suppressing class I histone deacetylases Thakur VS , Gupta K, Gupta S. Carcinogenesis.2012;33(2). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2024
Author Details