In silico Design of a Quadruplex RNA Aptamer Against Carcinoembryonic Antigen, and Evaluating its Potential as a Drug Delivery Vehicle
Download
Abstract
Background: Colorectal cancer is the third most common cancer worldwide, with carcinoembryonic antigen (CEA) being a key biomarker for tumor prognostic assessment. This study focused on the in silico development of an RNA aptamer capable of efficiently targeting the CEA biomarker, and evaluating its potential to act as a drug delivery vehicle of chemotherapeutic anticancer drugs.
Methods: In this study, five G-quadruplex RNA aptamers were manually designed and docked with the CEA protein. The aptamer sequence with the best docking score was optimized through various mutations, and these mutant sequences underwent drug-aptamer docking using AutoDock vina, followed by further docking experiments with CEA, with and without the drug, using the Hdock server. The best aptamer-drug configuration and the entire (aptamers-drug)-CEA complex were then analyzed using molecular dynamics (MD) simulation.
Results: Irinotecan exhibited the highest binding affinity with three mutant sequences compared to 5-FU and Raltitrexed, with the seq 3-5/Irinotecan complex showing the best configuration (score: -11.6). This complex also demonstrated a good binding affinity with the target CEA (HDOCK score: -393.07). Throughout 31.6 ns simulation, the RMSD plot of the (aptamer-drug)-CEA complex showed an average of 8.7 A°. The residues of interacting amino acids had lower RMSF than 1.4 Å, except for the ALA A0 (RMSF:1.74 Å). Rg plot showed minimal fluctuation, which were maintained between 19.2 Å and 20.2 Å. The mean SASA was in the range of 112.8–125.8 nm2. H-bonds were observed with an average of 7.3 hydrogen bonds. MD simulation results indicated that the (aptamers-drug)-CEA complex maintained a stable, low-flexibility, and relatively compact structure throughout the simulation.
Conclusion: The study concludes that the designed quadruplex RNA aptamer can potentially target the CEA biomarker with good binding affinity and serves as an effective carrier for Irinotecan. However, in vitro and in vivo experiments are necessary for further validation.
Introduction
Cancer remains a devastating disease with a rising mortality rate annually, presenting a significant global health challenge [1]. Among the various types of cancer, colorectal cancer (CRC) stands out as the third most common globally and a leading cause of cancer-related deaths [2]. A key biomarker utilized in CRC for tumor screening and prognostic assessment is Carcinoembryonic antigen (CEA) [2]. It is a glycoprotein with a molecular weight of 200 kDa, belonging to the CEACaM immunoglobulin family, and plays roles in cell proliferation, adhesion, and migration. In normal cells, CEA is primarily located in the endoluminal section of the cell membrane, where it inhibits cell apoptosis and tumor development [3]. The current treatment modalities for CRC predominantly encompass chemotherapy, radiation therapy, immunotherapy, and palliative care, with chemotherapy playing a pivotal role in managing the disease [4]. Several chemotherapy drugs have been approved for CRC treatment, such as irinotecan, leucovorin, oxaliplatin, capecitabine, and 5-fluorouracil [5]. However, the nonselective cytotoxic mechanism of chemotherapeutic agents often leads to significant side effects during and post-treatment. Therefore, targeted therapy has emerged as a promising systemic approach for CRC treatment [6], aiming to enhance drug efficacy while minimizing adverse effects. The selection of specific ligands that target tumor cells with no affinity for normal cells is the most widely used strategy in targeted therapy [7]. Aptamers are short single-stranded oligonucleotides typically ranging from 20-200 nucleotides in length. They form unique three-dimensional (3D) structures such as single-stranded segments, triplex, G-quadruplex, hairpin, internal loop, and bulge, which provide proper sites for binding to diverse targets including nucleic acids, proteins, whole cells, viruses, bacteria, and small molecules [8]. The systematic evolution of ligands by exponential enrichment (SELEX) method is commonly employed for aptamer development [9]. An alternative strategy to SELEX that has been suggested in the past decade and a half is computational methods of bioinformatics, such as docking and molecular dynamics, to design aptamers for many purposes [10].
This study focused on the in silico development of an RNA aptamer capable of efficiently binding to the cell surface of tumor cells targeting the CEA tumor marker, and evaluating its potential to act as a drug delivery vehicle. Molecular docking analyses were conducted to assess the binding affinity of the designed aptamer with various drugs, followed by molecular dynamics simulations to evaluate the binding stability of the best aptamer-drug configuration, and for the entire complex ((aptamers-drug)-CEA) complex.
Materials and Methods
Structure Retrieval
The crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) (PDB ID: 2QSQ, with a resolution of 1.95 Å) was retrieved from the RCSB Protein Data Bank (PDB). The three-dimensional (3D) structures of the drugs Irinotecan (CID: 60838), 5-Fluorouracil (CID: 3385), and Raltitrexed (CID: 135400182) were retrieved from the PubChem web server in simple document format (SDF).
Design of RNA aptamers, and prediction of the G-rich quadruplexes
Five aptamers were manually designed against the CEA protein. The aptamers were analyzed for the presence of quadruplex-forming- G-rich sequences (QGRS) by using the web-based QGRS Mapper server (QGRS Mapper | G-quadruplex analysis tool) with default settings; QGRS Max length of 36, min G-Group size of 2 and loop size of 0 to 36. Each predicted structure was given a G-score, with higher scores indicating greater stability of the G-quadruplex.
The primary RNA sequences were further examined for secondary structures (2D) prediction using the RNAfold web server 2.6.3 (http://rna.tbi.univie.ac.at/ cgi-bin/RNAWebSuite/RNAfold.cgi ), with the following options: minimum free energy (MFE) only, avoiding isolated base pairs, RNA parameters, rescale energy parameters to given temperature (37°C), and a salt concentration of (1.021 molar). The RNAfold provided MFE calculations and displayed the MFE structure in bracket notation. The RNA sequences and the dot-bracket 2D structure notations were input into the RNA Composer server (https://rnacomposer.cs.put.poznan.pl/) to generate 3D models.
The structural elements repository was configured with the following settings: generating A-RNA-based double helices and single strands was turned off, only X-ray determined structures were used with a resolution threshold of 3.0 Å. No atom distance or torsion angle restraints were introduced.
Subsequently, the 3D structures of the aptamers were saved as PDB files for use in the ligand (aptamer)- receptor (CEA) docking.
Prior to docking, water molecules, ions and small molecules (glycerol, and chloride ions) present in the CEA retrieved structure were removed using autodock tools V 1.5.7. The docking was performed using the HDOCK server (http://hdock.phys.hust.edu.cn/), and no binding site was identified. HDOCK is an online server designed for docking proteins with other proteins or with DNA/RNA. It employs a hybrid approach that combines template-based modeling with ab initio docking methods. This tool is crucial for investigating molecular interactions involving proteins and nucleic acids. The Huang Laboratory is responsible for its development.
Structural modification of the aptamer through different mutations
The best-bound aptamer with the highest affinity to the target was selected based on the docking score, minimum free energy, and G score. Subsequently, different types of mutations including deletion, replacement, and insertion were introduced to enhance binding affinity and achieve a more stable structure. The detailed interaction profile between the selected aptamer and the CEA target domain was analyzed using the Protein-Ligand Interaction Profiler (PLIP) web tool (https://plip-tool.biotec.tu-dresden.de/ plip-web/plip/index). This analysis provided insights into the specific amino acids and nucleotides involved in the interaction, including hydrogen bonds, π-stacking, hydrophobic interactions, and ionic interactions.
Drug-aptamer docking
Three aptamers demonstrating high affinity were chosen for drug-aptamer docking. The docking was performed using AutoDock vina and AutoDockTool. The 3D structures of the drugs Irinotecan, 5-Fluorouracil, and Raltitrexed were converted from SDF format to PDB format using OpenbableGUI V 2.4.1. Then, both the receptor (aptamer) and ligands (drugs) PDB structures were converted to PDBqt format. The receptors’ structures were modified by adding polar hydrogen atoms and Kollman charges using AutoDock Tool. Single bonds of drugs were allowed to rotate freely, whereas the aptamer remained rigid.
(Drug-aptamer)-target docking
Based on the docking scores, the drug exhibiting the highest binding affinity with the aptamers was selected for further docking experiments with the target CEA. The docking was performed using the HDOCK web server for aptamer-CEA complexes both in the presence of the drug ((drug-aptamer)- CEA complex) and in its absence.
Molecular dynamics (MD) simulation
The molecular dynamics simulation of the drug-aptamer and (drug-aptamer)- CEA complexes was performed using NAMD v3.0b4 (https://www.ks.uiuc.edu/Research/ namd). The initial configurations of the complexes were generated using the CHARMM-GUI Solution Builder feature (https://www.charmm-gui.org ) based on the provided PDB files. Each complex was placed in the center of a box with a 10 Å margin on all sides, hydrated with a TIP3P water model, and neutralized by adding 0.15 M NaCl ions. The simulations ran for about 31.6 ns using the CHARMM36m force field at a temperature of 300K and a pressure of 1 bar. The resulting trajectory data were analyzed using VMD v1.9.4a53. Key parameters such as root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), solvent-accessible surface area (SASA), radius of gyration (Rg), and hydrogen bond interactions (H-bonds) were determined.
Results
Aptamer design and optimization
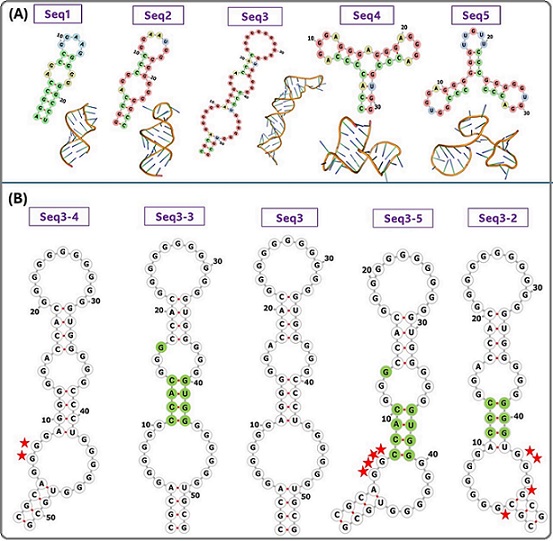
Five RNA aptamers were designed and docked with the CEA protein. The secondary structures of the designed aptamers are composed of G-quadruplex (G4), stems, hairpin loops, inner loops, and Watson–Crick base pairing, as shown in Figure 1A.
Figure 1. (A) The Secondary and Tertiary Structures of the RNA Aptamers (B) Examples of Mutations Introduced in seq-3 Aptamer. Red stars represent the deletion, and the green highlight represents the replacement.
The MFE, G score, and docking scores of the aptamer-CEA complexes are listed in Table 1.
Table 1. (A), Initial Sequences with Minimum Free Energy, G-score, and Docking Scores. (B), New sequences generated by introducing mutations in seq-3 aptamer, with minimum free energy, G-score, H- bonds and docking scores.
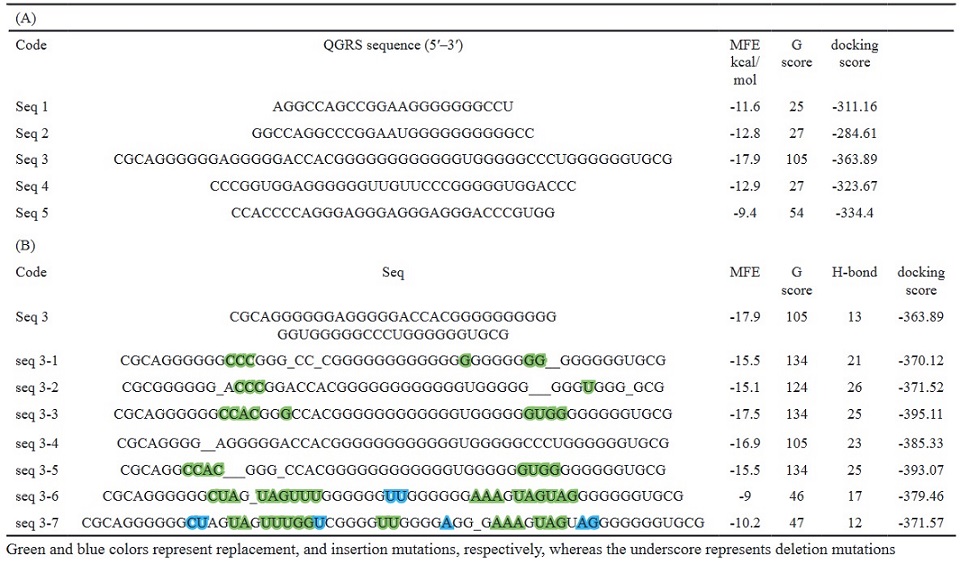
Green and blue colors represent replacement, and insertion mutations, respectively, whereas the underscore represents deletion mutations
Seq 3 with a length of 53-mer was the best-binding structure, exhibiting the highest G-score of 105, and the lowest docking score (score: -363.89), and MFE (17.90 kcal/mol). Therefore, it was optimized to obtain a more stable structure by introducing different mutations including deletion, replacement, and insertion. New mutant-generated sequences are listed in (Table 1B). As shown in (Table 1B), three structures (seq 3-3, seq 3-4, and seq 3-5) showed the lowest docking scores compared to the original structure, suggesting that the minor nucleotide variations can improve the stability of the aptamer and its binding properties. These three sequences were selected for drug-aptamer docking.
Molecular docking of drug-aptamer and (drug-aptamer)- target
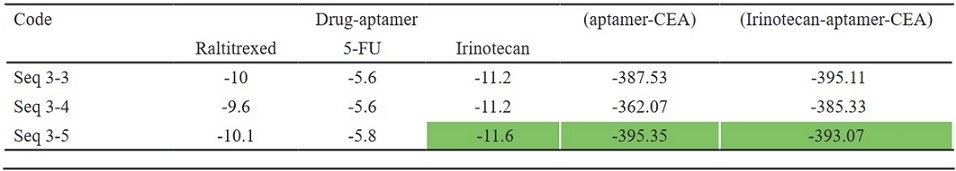
Based on the docking scores, number of H-bonds, and MFE values, three sequences (seq 3-3, seq 3-4, and seq 3-5) were chosen for drug-aptamer docking. The results are presented in Table 3. Irinotecan showed the highest binding affinity with all three aptamers compared to –FU and Raltitrexed, and the seq 3-5-Irinotecan complex was the best configuration (score: -11.6), so it was selected for further docking experiments with the target CEA. The docking was performed both in the presence of the drug ((drug-aptamer)- CEA complex) and in its absence (Table 2).
Table 2. Docking Scores of the Drug-aptamer, (aptamer-CEA), and (Irinotecan-aptamer-CEA) Complexes.
Molecular dynamics simulation of the aptamer-drug complex
MD simulation was performed for the best aptamer- drug configuration (seq 3-5/irinotecan complex) to evaluate its potential to act as a drug delivery vehicle.
The highest RMSD value was 11.83 A°, and the average RMSD was (7.8±1.49 A°). The RMSD graph of seq 3-5 aptamer/drug complex showed a fluctuating trend from 0 to 11.83 A° throughout the first 15.75 ns. Then it gradually declined with a clear tendency to stabilize. After 24.7 ns, the RMSD pattern stabilized with an average of (7.8 ±0.4 A°) until the end of the simulation (Figure 2 A).
Figure 2. RMSD Plot (A) and H Bond (B) of the seq3-5 Aptamer/drug Complex During the MD Simulation.
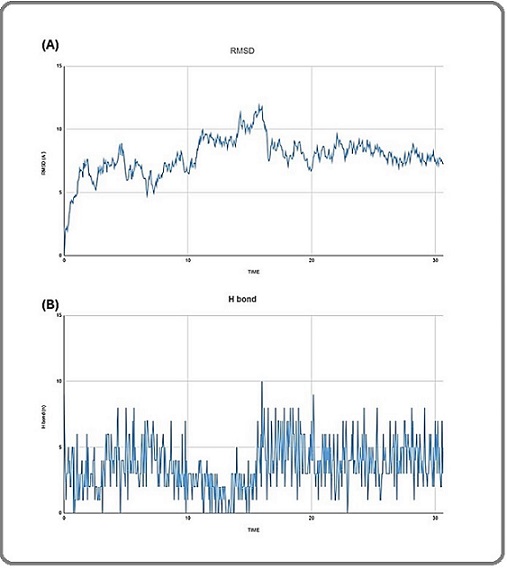
The total number of H-bonds that appeared in the MD simulation trajectory for the seq 3-5 aptamer/drug complex is shown in (Figure 2 B). The complex had an average of 3.6±1.87 hydrogen bonds, with the highest number of 10 bonds during the simulation. At 31.6 ns, the final number of hydrogen bonds was 5.
Molecular dynamics simulation of (aptamer-Drug)-CEA complex
Root Mean Square Deviation (RMSD)
Figure 3 A displays the RMSD graph of the seq 3-5 aptamer-drug/CEA complex.
Figure 3. RMSD plot (A), RMSF Plot (B), Rg Plot (C), and SASA Plot (D) of the (seq3-5 aptamer-Drug)/CEA Complex During the MD Simulation.
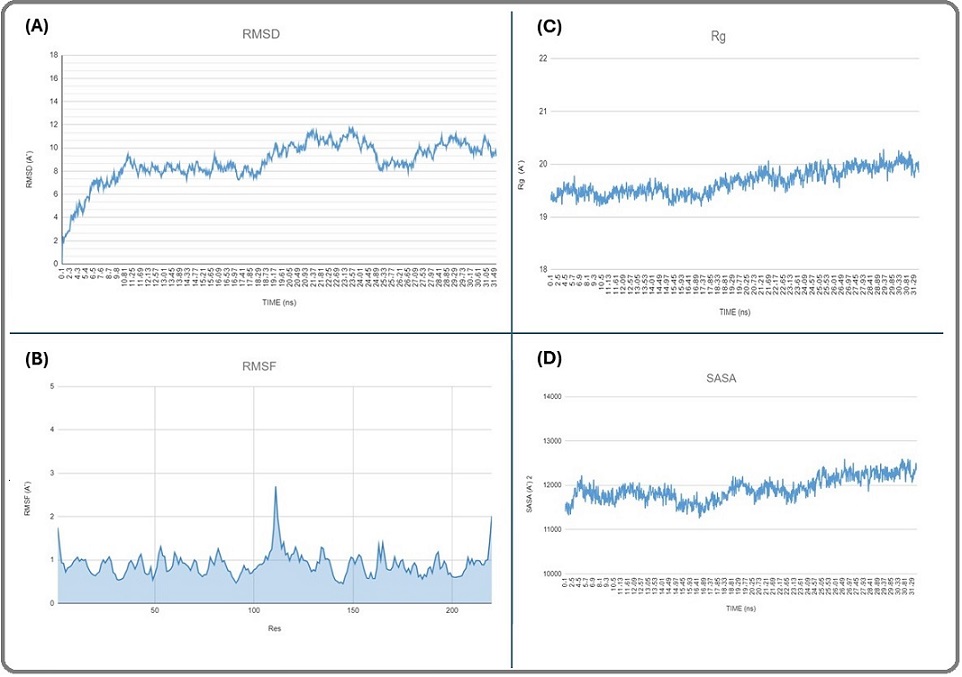
The highest RMSD value was 11.72 A, and the average RMSD was 8.7±1.7 A. The RMSD pattern showed an upward trend from 0 to 9.2 Å throughout the first 11,09 ns. Then, the complex reached a short-term steady state from 11.35 to 15.75 ns in the simulation, with an average of 8.1±0.3 A. Then it suddenly increased and showed several fluctuations, eventually stabilizing within the range of (9-11) Å until the end of the simulation.
Root Mean Square Fluctuation (RMSF)
The RMSF of the seq 3-5 aptamer-drug/CEA simulated complex was analyzed (Figure 3 B). The RMSF graph represents the residues on the x-axis and the fluctuation values on the y-axis. The highest and lowest RMSF values were 2.6 Å and 0.4 Å, respectively, and the average RMSF was 0.9±0.2 Å.
The radius of gyration (Rg)
Figure 3 C displays the Rg plot of the seq 3-5 aptamer-drug/ CEA complex. Rg-time fluctuations were maintained between 19.2 Å and 20.2 Å throughout 31.61 ns, with an average of (19.6±0.2) Å.
Solvent accessible surface area (SASA)
The surface area of the seq 3-5 aptamer-drug/ CEA complex was in the range of 112.8–125.8 nm2 with an average of 119.2 nm2 (Figure 3 D).
Hydrogen bonds (H-bond) analysis
The total number of H-bonds that appeared in the MD simulation trajectory for the seq 3-5 aptamer-drug/CEA complex is shown in (Figure 4 A).
Figure 4. (A), Number of Hydrogen Bonds of the (seq3-5 aptamer-Drug)/CEA complex during the 31.6 ns MD Simulation; (B), Visualization of the Hydrogen Bonds Formed by the seq3-5 Aptamer/drug and CEA Protein in the Last Frame of MD Simulation. The aptamer is colored in gray.
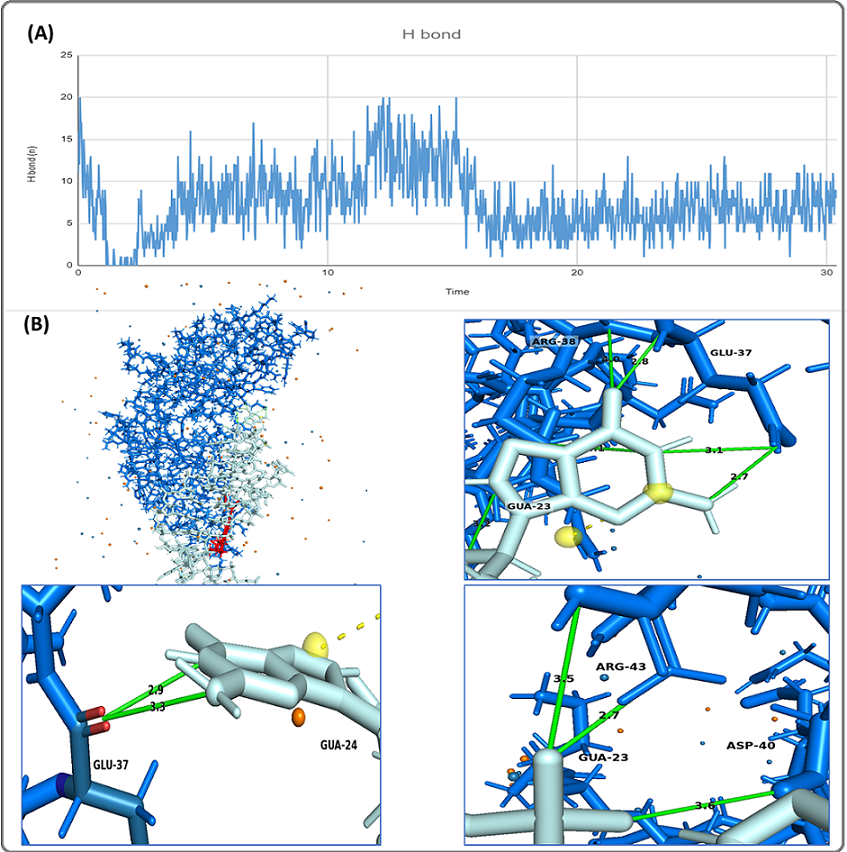
The complex had an average of 7.38±3.7 hydrogen bonds, with the highest number of 20 bonds at the beginning of the simulation. At 30.4 ns, the final number of hydrogen bonds between the CEA protein and seq3-5 aptamer-drug was 8. In the last frame of the MD simulation for the complex, hydrogen bonds appeared in GLU-37, ARG-28, ARG-43, ASP-40 (Figure 4 B).
Discussion
The utilization of computational methods to study biomolecular interactions has significantly advanced the analysis of biological systems, offering valuable insights for predicting biomolecule’s activity and understanding biological processes. In-silico methods are commonly employed in drug discovery, protein-protein interactions, aptamer-protein interactions, and more [11].
In the current study, G-quadruplex RNA aptamers targeting the CEA protein were designed. These aptamers have G-rich sequences capable of forming four-stranded structures that are crucial for ligand binding [12]. The 3D structures of the aptamers were then docked with the CEA protein.
The aptamer sequence with the best docking score was selected, and random mutations were introduced in its sequence to enhance stability and affinity. Subsequently, three aptamers demonstrating high affinity were chosen for drug-aptamer docking using AutoDock vina and AutoDockTool. Before docking, the receptors’ structures were modified by adding polar hydrogen atoms and Kollman charges using the AutoDock Tool.
Polar hydrogen atoms play a crucial role in hydrogen bonding interactions, leading to dipole‐dipole and other weak interactions, while non‐polar hydrogens contribute to van der Waals interactions, which do not have very significant importance. Kollman charges represent the template value for an amino acid, derived from the corresponding electrostatic potential [13].
Molecular docking aids in determining the optimal interaction between the ligand and target protein, estimating the energy released during the interaction. AutoDock, an open‐source automated docking program developed by the Molecular Graphics Lab at Scripps Research Institute, La Jolla, CA 92037, United States, is utilized for calculating interaction profiles of biological macromolecules like proteins/enzymes and ligands [13].
Among the tested drugs, Irinotecan exhibited the highest binding affinity with all three mutant sequences compared to 5-FU and Raltitrexed. In clinical practice, the treatment of CRC has been closely associated with the use of Irinotecan [4].
Irinotecan, a chemotherapeutic agent derived from camptothecin, targets DNA topoisomerase I (topo I). It is well established that the levels of topo I am upregulated in certain cancer types, and thus inhibiting of topo I activity can affect tumor cells’ proliferation [14]. Irinotecan and its active metabolite SN-38 form a ternary complex with Topo I-DNA. When this complex collides with the progressive replication fork, it will form double-stranded DNA unwinding. This process results in irreversible stagnation of the replication fork, cell death, and and a potent anti-tumor effect by interfering with the process of DNA replication in tumor cells [4].
Based on the docking scores, the seq 3-5-Irinotecan complex was the best configuration. Due to its guanine-rich nature (69%), the seq 3-5 aptamer was analyzed for the presence of QGRS. An analysis revealed 10703 potential structures for the seq 3-5 aptamer to form a G-quadruplex structure, with a maximum G-Score of 134, indicating a high likelihood of the seq 3-5 aptamer forming a G-quadruplex structure.
The seq 3-5-Irinotecan complex was selected for further docking experiments with the target CEA using the Hdock server. HDOCK is a web server designed for protein-protein and protein-DNA/RNA docking, utilizing a hybrid algorithm that combines template-based modeling and ab initio-free docking. It plays a crucial role in facilitating the exploration of molecular interactions between proteins and nucleic acids. HDOCK was developed by Huang Laboratory. Based on docking results, seq 3-5 aptamer was selected as a drug carrier for Irinotecan.
The stability of docked complexes and the binding pose obtained in molecular docking is widely verified by molecular dynamics simulation [15].
MD simulation was performed for the best aptamer-drug configuration (seq 3-5-irinotecan) to assess its potential as a drug delivery vehicle, and for the entire complex ((aptamer-drug)-CEA) complex to gain deeper insights into the stability, flexibility of the residues and the compactness of the entire complex. Parameters including RMSD, H-bonds, RMSF, SASA and Rg were determined. One of the critical parameters to analyze a protein-ligand complex is the RMSD of the protein backbone C-α atoms, which characterizes its overall conformational stability in a dynamic state during the simulation [16]. The RMSD plot showed an average of 7.8 A°, and 8.7 A° for the aptamer-drug, and the (aptamers-drug)-CEA complexes, respectively, indicating that the aptamer remained sufficiently bound to the drug and the target CEA throughout the simulations, resulting in a stable conformation for the entire complex.
RMSF analysis of a protein-ligand complex quantifies the degree of flexibility or mobility exhibited by individual atoms within a molecular system throughout a simulation [17]. Residues showing higher levels of RMSF imply increased flexibility, whereas lower RMSF fluctuations imply lesser flexibility [18]. The peaks in L_RMSF may indicate the presence of regions that are engaged in binding or unbinding processes or locations that exhibit heightened conformational dynamics [17]. It can be observed from the RMSF plot that some residues display large changes, particularly the residues (LEU A 110, PRO B 108, and ALA B 0), which showed RMSF values of 2.69 Å, 2.01 Å, 1.97 Å, respectively. These significant fluctuations of RMSF values were observed for the residues of amino acids not bound to the aptamer. The residues of interacting amino acids did not show significant fluctuations during the MD simulation, and had lower RMSF than 1.4 Å, except for the ALA A 0 (RMSF:1.74 Å), indicating stable and low flexibility of the complex.
Rg is a measure of the distribution of mass around the center of mass of the protein, providing insights into its structural properties and dynamics. By calculating the Rg at different time points during MD simulation, one can assess the expansion or contraction of the protein structure, as well as fluctuations in its shape [18]. Rg plot of the entire complex showed minimal fluctuation, which were maintained between 19.2 Å and 20.2 Å throughout 31.61 ns, indicating a relatively compact and stable structure throughout the simulation.
SASA was calculated to assess the exposed surface of the protein in the presence of solvent. It measures the fraction of the total surface area of the protein that is accessible to the solvent. Since external molecules are integrated with solvent, the surface area exposure to solvent means exposure to external molecules [19]. It is a crucial parameter in protein structure analysis as it provides insights into the protein’s folding, stability, and interactions with other molecules [18]. The high SASA values indicate structure expansion and increased solvent accessibility [20], while a lower SASA value represents a compact structure. An increase or decrease in SASA values represents changes in the protein’s structural conformation [21]. The mean SASA of the entire complex was in the range of 112.8–125.8 nm2, indicating a compact structure and that residues are less exposed to water, and the area of the binding interface was thus large.
Hydrogen bonding occurs when a weakly positive hydrogen atom in one molecule is attracted to a highly electronegative atom in another [22].
H-bonds were observed in the MD simulation trajectories, with an average of 3.6 and 7.3 hydrogen bonds for the aptamer-drug complex, and the entire complex, respectively, which indicates that the complexes had a relatively stable binding. For the entire complex, ARG38, GLU37, and ARG105 were the most important amino acids because their side chains could form hydrogen bonds with the aptamer with high occupancy rates (ARG38/ GUA24: 39.93%, ARG38/ GUA24: 38.95%, ARG38/ GUA23: 26.13%, GLU37/ GUA23: 36.89%, GLU37/ GUA24: 19.56%, GLU37/ GUA23: 10.60%, ARG105/ GUA27: 38.04%, ARG105/ GUA26: 10.27%). On the other hand, ARG105, VAL106, and GLY84 could form hydrogen bonds with the drug with high occupancy rates (ARG105 (main): 134.26%, ARG105 (side): 44.62%, VAL106 (main): 97.53%, GLY84 (main): 65.08%).
In the current study, a quadruplex RNA aptamer was designed through a non-SELEX in silico method. The designed aptamer showed a potential for targeting CEA biomarker with good binding affinity and serving as an effective carrier for Irinotecan chemotherapy drug.
A previous study [23] employed an in silico approach to screen the mutation sequences of reported DNA aptamers against carcinoembryonic antigen (CEA) using the online ZDOCK server. The top three ZDOCK scores for the analyzed DNA sequences were (P-ATG: 1535.11, GAC-P: 1466.01, and P-GTG: 1434.82). A higher ZDOCK score indicates higher binding ability. To compare these findings with our study, molecular docking was conducted for the seq 3-5 aptamer-CEA complex using the ZDOCK server (https://zdock.wenglab.org/), and the results revealed that the seq 3-5 aptamer-CEA complex yielded a higher docking score (ZDOCK score: 1545.99) than those reported in the previous study. In another study [24], DNA aptamers with affinity to CEA protein were also selected using SELEX method, but the study did not provide the full sequence of the aptamer, so we could not compare with it.
CEA is one of the most commonly utilized tumor markers in clinical diagnostics, primarily functioning to mediate the adhesion reaction between cancer cells and stromal collagen. It also plays a significant role in tumor growth and metastasis. In healthy individuals, serum CEA levels are typically low; however, elevated levels can be observed in various cancers, including those of the breast, colon, and lung [25]. Consequently, developing detection techniques for CEA holds substantial potential for early cancer diagnosis.
However, it is worth noting that in silico methods have their limitations. One such limitation of the current study is that MD simulations could only be performed over a period of 31.6 nanoseconds, due to the limited computing power.
Despite advancements in computer modeling and DNA/RNA folding algorithms, challenges related to computer processing load, especially with more flexible docking approaches, along with difficulties in accurately mimicking the electrochemical environment these molecules would encounter in vivo, indicate that predictions at this stage must be validated in vitro before any definitive conclusions can be drawn [26]. Despite the limitations, the in silico approach remains a valuable tool for enhancing our understanding of aptamer-protein interactions and offers promising strategies for the advancement of aptamer technology.
In conclusion, this study is the first to test the ability of a CEA aptamer to act as a drug delivery vehicle for chemotherapeutic anticancer drugs. We hope that this study will pave the way for the development of delivery systems based on cancer-specific CEA aptamers for the selective accumulation of the drugs in target tissues. This would enhance the selectivity of cancer chemotherapy, increase toxicity to cancer cells, while maintaining a high rate of viability for normal cells. However, further studies should be considered to evaluate the stability of the (Irinotecan-aptamer)/CEA complex in vitro and in vivo and to enhance its binding capacity.
Acknowledgments
Statement of Transparency and Principals
• Author declares no conflict of interest
• Study was approved by Research Ethic Committee of author affiliated Institute.
• Study’s data is available upon a reasonable request.
• All authors have contributed to implementation of this research.
References
- Colorectal cancer: A comprehensive review based on the novel drug delivery systems approach and its management Hani U, Honnavalli y, Yasmin Begum M., Yasmin S, Osmani R, Ansari M. Journal of Drug Delivery Science and Technology.2021;63. CrossRef
- Postoperative carcinoembryonic antigen (CEA) levels predict outcomes after resection of colorectal cancer in patients with normal preoperative CEA levels Zhang C, Zheng W, Lv Y, Shan L, Xu D, Pan Y, Zhu H, Qi H. Translational Cancer Research.2020;9(1). CrossRef
- Significance of carcinoembryonic antigen detection in the early diagnosis of colorectal cancer: A systematic review and meta-analysis Wang R, Wang Q, Li P. World Journal of Gastrointestinal Surgery.2023;15(12). CrossRef
- The effective combination therapies with irinotecan for colorectal cancer Chai Y, Liu J, Zhang S, Li N, Xu D, Liu W, Fu R, Tang Y. Frontiers in Pharmacology.2024;15. CrossRef
- Application of smart nanoparticles as a potential platform for effective colorectal cancer therapy | Request PDF Iranpour S, Bahrami AR , Saljooghi ASh , Matin MM . ResearchGate.. CrossRef
- Promising Therapeutic Strategies for Colorectal Cancer Treatment Based on Nanomaterials Krasteva N, Georgieva M. Pharmaceutics.2022;14(6). CrossRef
- Advances in Screening and Development of Therapeutic Aptamers Against Cancer Cells Li Z, Fu X, Huang J, Zeng P, Huang Y, Chen X, Liang C. Frontiers in Cell and Developmental Biology.2021;9. CrossRef
- Anti-Proliferative Effect of Doxorubicin-Loaded AS1411 Aptamer on Colorectal Cancer Cell Lohlamoh W, Soontornworajit B, Rotkrua P. Asian Pacific journal of cancer prevention: APJCP.2021;22(7). CrossRef
- Aptamers as an approach to targeted cancer therapy Mahmoudian F, Ahmari A, Shabani S, Sadeghi B, Fahimirad S, Fattahi F. Cancer Cell International.2024;24(1). CrossRef
- Methods and Applications of In Silico Aptamer Design and Modeling Buglak AA , Samokhvalov AV , Zherdev AV , Dzantiev BB . International Journal of Molecular Sciences.2020;21(22). CrossRef
- In-Silico Selection of Aptamer Targeting SARS-CoV-2 Spike Protein Lin Y, Chen W, Hwu E, Hu W. International Journal of Molecular Sciences.2022;23(10). CrossRef
- In silico design of quadruplex aptamers against the spike protein of SARS-CoV-2 Behbahani M, Mohabatkar H, Hosseini B. Informatics in Medicine Unlocked.2021;26. CrossRef
- Identification of Severe Acute Respiratory Syndrome Coronavirus-2 inhibitors through in silico structure-based virtual screening and molecular interaction studies Azad I, Khan T, Maurya AK , Irfan Azad M, Mishra N, Alanazi AM . Journal of molecular recognition: JMR.2021;34(10). CrossRef
- Establishment and Characterization of an Irinotecan-Resistant Human Colon Cancer Cell Line Wu Z, Yang Y, Zeng L, Patel H, Bo L, Lin L, Chen Z. Frontiers in Oncology.2020;10. CrossRef
- Molecular docking and dynamics studies on propolis sulabiroin-A as a potential inhibitor of SARS-CoV-2 Fatriansyah JF , Rizqillah RK , Yandi MY , Fadilah N, Sahlan M. Journal of King Saud University. Science.2022;34(1). CrossRef
- An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2 Sharma J, Kumar Bhardwaj V, Singh R, Rajendran V, Purohit R, Kumar S. Food Chemistry.2021;346. CrossRef
- Molecular Docking and Structure‐Activity Relationship Analysis of Target Compounds against Glyceraldehyde‐3‐Phosphate Dehydrogenase in Azithromycin‐Resistant Neisseria gonorrhoeae Yakobi S, Zuma L, Pooe O. ChemistrySelect.2024;9. CrossRef
- Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach Bagewadi ZK , Yunus Khan T. M., Gangadharappa B, Kamalapurkar A, Mohamed Shamsudeen S, Yaraguppi DA . Saudi Journal of Biological Sciences.2023;30(9). CrossRef
- Drug repurposing for SARS-CoV-2: a high-throughput molecular docking, molecular dynamics, machine learning, and DFT study Kashyap J, Datta D. Journal of Materials Science.2022;57(23). CrossRef
- Identification of PICK1 PDZ-Domain Antagonists: Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulation Analyses Rathod SB . ChemRxiv.2021. CrossRef
- Revisiting the inhibitory potential of protein kinase inhibitors against NEK7 protein via comprehensive computational investigations Ejaz SA , Aziz M, Zafar Z, Akhtar N, Ogaly HA . Scientific Reports.2023;13(1). CrossRef
- Molecular Dynamics Simulation of Lenalidomide Interaction with CRBN Protein: A target for immunomodulatory Drugs Esmaeili A, Yoosefian M, Mahani M. South African Journal of Chemistry.2023;77. CrossRef
- In silico post-SELEX screening and experimental characterizations for acquisition of high affinity DNA aptamers against carcinoembryonic antigen Wang Q, Cui H, Du J, Lv Q, Song X. RSC Advances.2019;9. CrossRef
- Development of radiopharmaceuticals based on aptamers: selection and characterization of DNA aptamers for CEA. INAC 2011: International nuclear atlantic conference Nuclear energy: new jobs for a better life, Brazil Correa CR , Andrade ASR , Augusto-Pinto L , Goes AM . 2011.
- Simultaneous Detection of VEGF and CEA by Time-Resolved Chemiluminescence Enzyme-Linked Aptamer Assay Man J, Dong J, Wang Y, He L, Yu S, Yu F, Wang J, et al . International Journal of Nanomedicine.2020;15. CrossRef
- Development and validation of anti-human Alpha synuclein DNA aptamer using computer modelling techniques-an in silico study Rock M, Zouganelis GD , Andrade AFB , Drake SJ , Alexiou A, Albrakati A, Batiha GES , Illingworth TA . Journal of Integrative Neuroscience.2022;21(1). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2024
Author Details