Diagnostic Values of Ki67, Cox2, CD31, E-cadherin, VEGF, MMP2, and MMP9 Protein Expression in Human Melanoma
Download
Abstract
Background: Cutaneous melanoma, the most severe form of skin cancer, has an unpredictable behavior and a high mortality rate. Recent research has identified new biomarkers and their associations with certain histopathological features, offering essential insights into diagnosis, prognosis, and therapeutic strategies.
Material and method: In this meticulously designed and executed study, we analyzed 85 formalin-fixed, paraffin-embedded (FFPE) tissue samples using the gold standard technique of immunohistochemistry (IHC) to characterize the protein expression profiles. We used a comprehensive panel of Ki67, Cox2, CD31, VEGF, MMP2, MMP9, and E-cadherin antibodies. Clinical stages were meticulously determined according to TNM classification, the thickness of the Breslow depth, and Clark levels. We employed robust statistical methods such as one-way ANOVA and curve estimation regression to analyze the data. The STRING database, a trusted resource, was used to identify protein-protein interactions and related pathways.
Results: The results of our study unveiled novel pairwise positive correlations between CD31 and Ki67 (r = 0.72), and between VEGF and Ki67 protein expression (r = 0.63). We also found a significant inverse relationship between the expression of E-cadherin and some biomarkers, such as Ki67, CD31, and VEGF (p < 0.001 for all). Additionally, our findings revealed a strong positive association between the extent of tumor invasion and the expression levels of Ki67, CD31, and VEGF. The overexpression of these proteins was significantly correlated with advanced clinical stages (p < 0.001 for all).
Conclusion: Our study suggests that the biomarkers we examined could be reliable potential diagnostic and prognostic biomarkers for cutaneous melanoma. These findings have the potential to significantly impact the diagnosis, prognosis, and treatment of this deadly disease, offering new avenues for improved patient care.
Introduction
Cutaneous melanoma is the most severe form of skin cancer, accounting for approximately one in five skin cancers, mainly in fair-skinned populations [1]. Melanoma is a malignant tumor predominantly known as one of the most lethal and resistant human tumors. Although the highest incidence is reported in Australia and New Zealand, the incidence is increasing significantly worldwide [2]. In 2020, approximately 325,000 new cases of melanoma were diagnosed worldwide, and nearly 57,000 people died from the disease [1]. Primary tumor thickness and ulceration are critical pathologic features determining survival and T-stage in cutaneous melanoma [3]. These features may not accurately assess tumor behavior or disease outcome; therefore, biomarkers may be significant in early diagnosis, prediction, and even response to treatment protocols [4].
Cadherins, as calcium-dependent transmembrane glycoproteins, mediate cell-cell adhesion. Epithelial cadherin (E-cadherin), a member of the cadherin family, is prominently expressed in the epidermis, except in the confined layer [5, 6]. Expression of E-cadherin protects the skin structure against the invasion of melanoma cells into the dermis through the down-regulation of invasion-related adhesion receptors and induction of apoptosis [5, 6]. In contrast, cyclooxygenase2 (Cox2), a key enzyme in the conversion of arachidonic acid to prostaglandin E (PGE), plays a detrimental role [7]. It enhances carcinogenesis, prevents cell apoptosis, and promotes tumor growth, angiogenesis, inflammation, tumor invasion, and metastasis [7, 8]. Importantly, Cox2 overexpression is strongly associated with melanoma progression [9], highlighting its potential implications.
The matrix metalloproteinase (MMP) family, including MMP2 and MMP9, contributes to the degradation of the extracellular matrix (ECM). The tissue inhibitory metalloproteinase (TIMP) acts as a counterbalance, inhibiting the activities of MMPs. However, an expression imbalance between MMP and TIMP can lead to accelerated tumor growth and invasion [10]. On the other hand, Ki67, expressed in all cell cycle phases except for the resting phase (G0), is a cause for concern. Excessive expression of Ki67 is associated with increased tumor cell proliferation, greater invasion, and rapid tumor growth, which can lead to disease progression, poor prognosis, and poor overall survival [11, 12].
In the context of angiogenesis in cutaneous melanoma, it is crucial to note the role of the vascular endothelial growth factor (VEGF) pathway. VEGF binds to VEGF receptor-1 (VEGFR1)/fms-like tyrosine kinase (Flt-1), and VEGFR2/kinase insert domain receptor (KDR)/ fetal liver kinase 1, all of which are highly expressed in vascular endothelial [13]. This pathway plays a significant role in neovascularization, which is measured using microvascular density (MVD). CD31 and CD34, two endothelial cell markers, are used to evaluate MVD. CD31 (PECAM-1; platelet/endothelial cell adhesion molecule-1) is a single-chain type 1 transmembrane protein involved in adhesive interactions between adjacent endothelial cells and between leukocytes and endothelial cells [14].
The primary objective of this study is to examine the expression levels of selected biomarkers related to differentiation, proliferation, angiogenesis, and cell migration in cutaneous melanoma, as illustrated in Figure 1.
Figure 1. Gene Ontology Shows the Biological Process of Selected Proteins. The STRING database was used to find any protein interactions between the selected proteins. Colored dots represent the involved pathways in cancers .
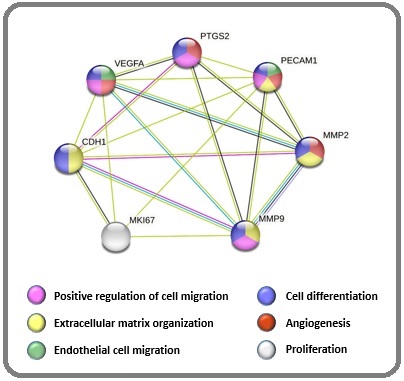
We also investigate their association with histopathological features such as TNM classification, the thickness of the Breslow depth and Clark levels. To accomplish this, we utilize immunohistochemistry (IHC), a widely accepted and well-documented method that allows us to characterize protein expression patterns while preserving tissue and cellular architecture.
Materials and Methods
Tissue collection and tumor specimens
To evaluate the expression of selected biomarkers and their relevant histopathological features, we conducted a cross-sectional study on patients diagnosed with cutaneous melanoma at the Cancer Institute of Iran, Imam Khomeini Hospital Complex, from 2010 to 2019. A total of 85 patients primarily diagnosed with cutaneous melanoma were included in our study. We excluded patients pathologically diagnosed with mucosal or uveal melanoma and the presence of metastasis. Demographic and histopathological characteristics such as gender, age, tumor location, tumor size, histopathological types, Clark levels, and Breslow thickness were available for this study (Table 1).
| Variable | No. | Frequency (%) |
| Gender | ||
| Female | 38 | 44.71 |
| Male | 47 | 55.29 |
| Age (years old) | ||
| <19 | 1 | 1.18 |
| 20-29 | 1 | 1.18 |
| 30-39 | 6 | 7.05 |
| 40-49 | 10 | 11.76 |
| 50-59 | 20 | 23.53 |
| 60-69 | 20 | 23.53 |
| 70-79 | 15 | 17.65 |
| ≥80 | 12 | 14.12 |
| Tumor site | ||
| Head & Neck | 19 | 22.35 |
| Lower Extremity | 55 | 64.71 |
| Trunk | 2 | 2.35 |
| Upper Extremity | 9 | 10.59 |
| Tumor size (cm) | ||
| <1 | 8 | 9.41 |
| 1 – 2.9 | 48 | 56.47 |
| 3 - 4.9 | 17 | 20 |
| ≥5 | 11 | 12.94 |
| Not available | 1 | 1.18 |
| Histological type | ||
| Acral lentigo | 27 | 31.77 |
| Lentigo maligna | 12 | 14.12 |
| Nodular | 33 | 38.82 |
| Superficial spreading | 3 | 3.53 |
| Not available | 10 | 11.76 |
| Clarck levels | ||
| I (in situ) | 13 | 15.29 |
| II | 15 | 17.65 |
| III | 19 | 22.35 |
| IV | 23 | 27.06 |
| V | 15 | 17.65 |
| Breslow’ depth thickness (mm) | ||
| In situ | 12 | 14.12 |
| ≤ 0.75 mm in thickness | 12 | 14.12 |
| > 0.75 – 1.5 mm in thickness | 7 | 8.23 |
| > 1.5 – 4 mm in thickness | 23 | 27.06 |
| > 4 mm in thickness | 31 | 36.47 |
According to the American Joint Committee on Cancer (AJCC), clinical stages were classified based on TNM classification, the thickness of Breslow depth, and Clarck levels, as detailed in Table 2.
| Clinical stage | TNM classification | Breslow depth (mm)/Clarck level | No. | Frequency (%) |
| 0 | pTis N0 M0 | In situ melanoma, Clarck level I | 12 | 14.12 |
| I | pT1 N0 M0 | ≤ 0.75 mm in thickness, Clarck level II | 19 | 22.35 |
| pT2 N0 M0 | > 0.75 – 1.5 mm in thickness, Clarck level III | |||
| II | pT3 N0 M0 | > 1.5 – 4 mm in thickness, Clarck level IV | 23 | 27.06 |
| III | pT4 N0 M0 | > 4 mm in thickness, Clarck level V | 31 | 36.47 |
| IV | Any pT Any N M1 | Distant metastasis | 0 | 0 |
Immunohistochemistry (IHC) study
Eighty-five formalin-fixed, paraffin-embedded (FFPE) tissue samples primarily diagnosed with cutaneous melanoma were obtained from the pathology department archives. The tissue array was prepared from the paraffin blocks containing the cancerous areas. Tissue array slides were first placed in the oven for 40 minutes and immediately immersed in xylene to remove the residual paraffin, then hydrated by an alcohol gradient. Slides were boiled in citrate buffer (pH=6.0) at 10 to 20% reduced power for 20 minutes to retrieve the antigens and incubated at room temperature for 30 minutes. After exhausting endogenous peroxidase using H2O2 in methanol for 15 min, sections were rinsed thrice with phosphate-buffered saline (PBS) and then blocked with 5% bovine serum albumin (BSA) for one hour at room temperature. Sections were rinsed thrice in PBS, incubated with specific antibodies for one hour at room temperature, and rinsed again thrice in PBS. Sections were incubated in horseradish peroxidase (HRP) according to instructions, rinsed thrice with PBS, and counterstained with Mayer’s hematoxylin method. Antibodies against Ki67 (rabbit monoclonal, SKU: 325, ready-to-use) and Cox2 (rabbit monoclonal, SKU: 306, dilution 1:50) were purchased from Biocare Medical (CA, USA). E-Cadherin (rabbit polyclonal, orb213706, dilution 1:100), VEGF (rabbit polyclonal, orb11554, dilution 1:100), CD31 (mouse monoclonal, orb181542, dilution 1:100), MMP2 (mouse monoclonal, (8B4): sc-13595, dilution 1:100) and MMP9 (rabbit polyclonal, orb11064, dilution 1:100) antibodies were obtained from antibodies- online GmbH (Aachen, Germany).
Immunoreactivity scoring
As shown in Figure 2, an experienced pathologist reviewed the sections.
Figure 2. Representative Images of Ki67, VEGF, Cox2, MMP2, CD31and MMP9 Staining in Melanomas by Immunohistochemistry (IHC). Cutaneous melanomas with low Ki67 (A), high Ki67 (B), low VEGF (C), high VEGF (D), Cox2-negative (E), Cox2-strong (F), MMP2-negative (G), MMP2-strong (H), low CD31 (M), high CD31 (N), MMP9-negative (O), and MMP9-strong (P) staining. VEGF and CD34 protein expression levels are used to assess microvessel density (MVD). Scale bars are equal to 100 μm .
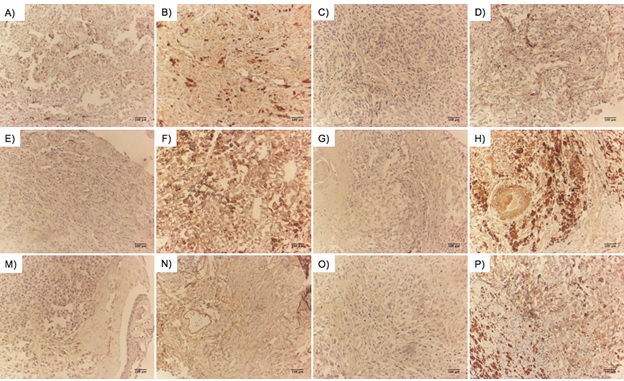
We calculated the microvessel density (MVD) as a measure to assess the expression levels of CD31 and VEGF. For this, we first found the hotspots in low-power fields and then measured the total scores of four intratumoral hotspots with high-power fields (HPFs). Additionally, for Ki67, we calculated the proliferation index (PI) based on the percentage of staining among one hundred malignant nuclei. Moreover, for Cox2, MMP2, and MMP9, we used the modified Allred score, which is a combination of two parameters including proportion and intensity scores, where a score of 0-1 was considered negative, 2-3 as low, 4-5 as moderate, and 6 to 8 as strong expression. For E-cadherin, we used the Allred score with few exceptions, where a score of < 3 was considered an abnormal change at low levels of transmembrane protein expression, 4 to 5 as intermediate levels, and 6 to 8 as negative with intact protein expression.
Statistical analyses
One-way ANOVA was calculated to compare a continuous variable with categorical explanatory variables. All values were expressed as mean and standard deviation (±SD). Curve estimation regression was fitted with the continuous variables, including Ki67, CD31, and VEGF protein expression. In all statistical analyses, p < 0.05 was considered significant. The RStudio statistical software version 1.2.5033 was used for statistical analysis. The STRING database was used to identify protein-protein interactions and related pathways [15].
Results
Clinical features and protein-protein interactions
Approximately half of the patients were male (55.29%), and the remaining half were female (44.71%). The age range of the patients was 15 to 94 years, with a median age of 65 years. Breslow’s depth thickness ranged from 0.15 to 35 mm, with a mean and standard deviation (±SD) of 11.13 and 18.37 mm. The distribution of tumor sites in the body was as follows: 19 (22.35%) tumors in the head and neck, 9 (10.59%) in the upper extremity, 2 (2.35%) in the trunk, and 55 (64.71%) in the lower extremity. The classified tumor sizes were: <1 cm (9.41%), 1 – 2.9 cm (56.47%), 3 – 4.9 cm (20%), and ≥ 5 cm (12.94%). Furthermore, the STRING database analysis, a robust research tool, for selected proteins exhibited the protein-protein interactions and related pathways. The selected proteins were involved in proliferation, angiogenesis, cell migration, cell differentiation, and extracellular matrix organization (Figure 1).
Association between selected biomarkers in cutaneous melanomas
Based on the data presented in Table 3, our results indicated a positive association between Cox2 protein expression and increased Ki67, CD31, and VEGF staining (p < 0.001 for all).
| Variable | Cox2 Protein Expression (mean ± SD) | p-value | |||
| Negative | Weak | Moderate | Strong | (among groups) | |
| Ki67 Expression | 5.88 ± 4.43 | 8.92 ± 4.67 | 11.31 ± 5.13 | 15.09 ± 4.3 | < 0.001 |
| CD31 for MVD | 8.76 ± 4.38 | 12.04 ± 6.69 | 14.81 ± 4.09 | 19.72 ± 3.44 | < 0.001 |
| VEGF for MVD | 10.65 ± 5.61 | 13.96 ± 6.1 | 16.66 ± 4.48 | 23.36 ± 3.56 | < 0.001 |
| MMP2 Protein Expression (mean ± SD) | |||||
| Negative | Weak | Moderate | Strong | ||
| Ki67 Expression | 5.07 ± 3.71 | 7.09 ± 3.95 | 11.57 ± 5 | 16.2 ± 3.73 | < 0.001 |
| CD31 for MVD | 9.13 ± 6.22 | 11.24 ± 4.57 | 14.69 ± 5.17 | 20 ± 3.71 | < 0.001 |
| VEGF for MVD | 10.8 ± 7.22 | 13.29 ± 4.46 | 16.85 ± 5.35 | 22.78 ± 3.53 | < 0.001 |
| MMP9 Protein Expression (mean ± SD) | |||||
| Negative | Weak | Moderate | Strong | ||
| Ki67 Expression | 6.88 ± 5.11 | 8.39 ± 5.25 | 11.19 ± 4.67 | 17 ± 3.21 | < 0.001 |
| CD31 for MVD | 10.35 ± 6.04 | 11.35 ± 5.69 | 15.42 ± 4.9 | 18.14 ± 5.46 | < 0.001 |
| VEGF for MVD | 11.94 ± 6.8 | 12.74 ± 5.14 | 18.19 ± 4.97 | 19.43 ± 7.68 | < 0.001 |
One-way ANOVA was applied to compute the p values among different Cox2, MMP2, and MMP9 protein expression levels of 85 melanomas. All variables displayed significant differences (p<0.05). Abbreviation; SD: standard deviation; MVD, microvessel density.
Furthermore, the overexpression of MMP2 was significantly correlated with higher expression levels of specific biomarkers, including Ki67, CD31, and VEGF (p < 0.001 for all). Similar findings were observed for the relationship between the expression levels of MMP9 and the intensity of Ki67, CD31, and VEGF staining (p < 0.001 for all). Conversely, the analysis of E-cadherin exhibited a significant inverse relationship between the expression of this protein and several biomarkers such as Ki67, CD31, and VEGF (p < 0.001 for all, Table 4).
| Variable | E-cadherin Protein Expression (mean ± SD) | p-value | ||
| Normal expression | Intermediate expression | No expression | (among groups) | |
| Ki67 Expression | 4.1 ± 3.35 | 10.52 ± 3.9 | 15.94 ± 4.11 | < 0.001 |
| CD31 for MVD | 8.61 ± 6.15 | 13.78 ± 4.63 | 18.18 ± 5.29 | < 0.001 |
| VEGF for MVD | 10.32 ± 6.12 | 16.17 ± 5.05 | 19.71 ± 6.43 | < 0.001 |
One-way ANOVA was applied to compute the p values among different E-cadherin protein expressions of 85 melanomas. All variables displayed significant differences (p<0.05). Abbreviation; SD: standard deviation, MVD: microvessel density.
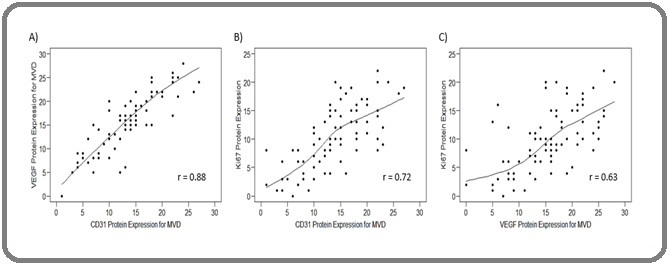
Also, a strong positive correlation was observed between CD31 and VEGF protein expression levels (r = 0.88, Figure 3A). Additionally, we found strong positive pairwise correlations between CD31 and Ki67 (r = 0.72, Figure 3B) and between VEGF and Ki67 protein expression (r = 0.63, Figure 3C).
Figure 3. Scatter Plots Demonstrate Correlations between Particular Biomarkers. Scatter plots illustrate the correlation between VEGF and CD31 expression (A), between Ki67 and CD31 expression (B), and between Ki67 and VEGF expression (C), indicating r > 0.7 as a strong correlation.
Clinical stages and their relationship with the protein expression patterns
The clinical stages were determined based on TNM classification, Breslow’s depth thickness, and Clarck levels, as described in Table 2. The classification showed that 12 (14.12%) patients were in stage 0, 19 (22.32%) patients in stage I, 23 (27.06%) patients in stage II, and 31 (36.47%) in stage III. The results revealed a significant association between different clinical stages and the protein expression levels of Ki67, CD31, and VEGF (p < 0.001 for all, Table 5).
| Variable | Clinical stage (mean ± SD) | p-value | |||
| 0 (in situ) | I | II | III | (among groups) | |
| Ki67 Expression | 2.83 ± 2.33 | 9.32 ± 4.63 | 10.87 ± 4.20 | 12.58 ± 5.20 | < 0.001 |
| CD31 for MVD | 6.92 ± 4.10 | 12.68 ± 5.20 | 13.83 ± 5.07 | 16.13 ± 5.68 | < 0.001 |
| VEGF for MVD | 9.17 ± 4.65 | 14.84 ± 4.52 | 15.70 ± 6.30 | 18.29 ± 6.11 | < 0.001 |
One-way ANOVA was applied to compute the p values among different Clark levels of 85 melanomas. All variables displayed significant differ- ences (p<0.05). Abbreviation; SD, standard deviation; MVD, microvessel density.
This association highlights the value of understanding the protein expression patterns in different tumor stages. Importantly, in advanced tumor stages, the protein expression levels of Ki67, CD31, and VEGF were notably high compared with early stages of tumors, underscoring the severity of these stages.
Discussion
Given the significance of the co-expression of biomarkers in predicting tumor progression and metastasis, to our knowledge, only a few publications have attempted to achieve this goal in different cancer types [12, 16, 17]. Therefore, this approach could be fruitful in further research on various cancer types, including melanomas. Here, we aimed to examine the protein expression levels of some critical biomarkers in cutaneous melanoma samples, including Ki67, Cox2, CD31, E-cadherin, VEGF, MMP2, and MMP9, and to correlate these biomarkers with specific clinicopathological features.
Several studies have indicated that the elevated Ki67 protein expression is strongly associated with increased Breslow thickness [18-20]. In this regard, our results were consistent with studies in which ki67 was directly related to Breslow thickness, which could be a marker for proliferation and invasiveness. However, Kamyab-Hesari et al. failed to demonstrate an association between ki67 expression and Breslow thickness [21].
Findings obtained through studies indicate that VEGF is up-regulated during melanoma progression and spread, and tumor-infiltrating cells expressing VEGF may contribute to the progression of melanoma [22]. Furthermore, Vesna Gajanin et al. demonstrated that tumors with high VEGF expression levels were associated with higher Breslow thickness [23]. Moreover, in a study of 60 melanoma cases, CD31 expression was correlated with the morphologic phases of melanocytic tumor progression with Clark’s levels of dermal infiltration and their associated metastatic potential [14]. In line with these, the highest protein expression of VEGF and CD31 protein expression was observed among tumors having higher clinical stages with higher Clarck levels and Breslow thickness. Additionally, Rajabi et al. described that the VEGF index was correlated with the Clark levels, but this was not observed for the distribution and intensity of VEGF [24]. However, some studies could not find any relationship between VEGF-C expression and Clark’s level [25, 26].
Further analysis demonstrating a significant reverse association between E-cadherin and Ki67 protein expression levels could indicate the promotion of epithelial-mesenchymal transition (EMT) and tumor progression. Similarly, the same results were observed between CD31 and E-cadherin and between VEGF and E-cadherin protein expression levels, activating angiogenic pathways and tumor invasion. However, we needed help accessing relevant articles showing the co-expression of these biomarkers in cutaneous melanoma to assess the validity of our data in comparison with related research. Our data showing a positive correlation between Ki67 with CD31 and VEGF protein expression may predict tumor progression. Additionally, the strong correlation between MMP2 with Ki67, CD31, and VEGF and between MMP9 with Ki67, CD31, and VEGF protein expression can predict tumor progression, invasion, and metastasis in cutaneous melanoma.
This study, while promising, has potential limitations such as a small sample size and the need for additional survival data. However, the potential of the molecular biomarkers investigated in this study in diagnosing cutaneous melanoma is significant. Analyzing a larger sample size with functional or genetic analysis of the tumor could provide further insights into the molecular mechanisms and pathways of the disease, thereby enhancing the prospects of our research. These biomarkers could serve as reliable diagnostic factors and predictors of cutaneous melanoma, potentially leading to the development of new targeted therapies and improved survival rates for melanoma patients. Further studies with a larger sample size are necessary to examine biomarker-biomarker and biomarker-clinical factor interactions in their prognostic impact on melanoma. These studies should prioritize early-stage patients most likely to benefit from personalized medicine with targeted therapies.
Abbreviations
Formalin-fixed paraffin-embedded: FFPE; Immunohistochemistry: IHC; Cyclooxygenase2: Cox2; prostaglandin E: PGE; Cluster of differentiation31: CD31; Matrix metalloproteinase2: MMP2; Matrix metalloproteinase9: MMP9; Epithelial-cadherin: E-cadherin; Vascular endothelial growth factor: VEGF; Extracellular matrix: ECM; Tissue inhibitor metalloproteinase: TIMP; Microvessel density: MVD; Tumor Node Metastasis: TNM; Epithermal growth factor receptor: EGFR; Platelet/endothelial cell adhesion molecule-1: PECAM-1; American Joint committee on Cancer: AJCC; Phosphate-buffered saline: PBS; Bovine serum albumin: BSA; Horseradish peroxidase: HRP; High-power field: HPF; Proliferation index: PI; Standard deviation: SD; Epithelial-mesenchymal transition: EMT; Centimeter: cm; Millimeter: mm; Micrometer: μM.
Funding
This project was supported by the Cancer Institute of Iran (Grant No: CRC48856).
Acknowledgments
We thank all patients included in this study. Biological material was taken from the IRAN NATIONAL TUMOR BANK, which is funded by the Cancer Institute of Iran. The Cancer Biology Research Center supported this work.
Authors’ contributions
NKY, AM, MR, AG, and AR designed the study. AM performed the IHC. AM and AG examined the IHC slides. NKY and MR collected the samples and the clinical data of patients. NKY and AR conducted the statistical analysis. NKY, AM, and AR drafted the manuscript, and all authors revised and approved it. AM and AR supervised the study. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040 Arnold M, Singh D, Laversanne M, Vignat J, Vaccarella S, Meheus F, Cust AE , et al . JAMA dermatology.2022;158(5). CrossRef
- Melanoma in Iran: a Retrospective 10-Year Study Ferdosi S, Saffari M, Eskandarieh S, Raziyeh R, Moghaddam MG , Ghanadan A, Shirkoohi R. Asian Pacific journal of cancer prevention: APJCP.2016;17(6).
- The eighth edition American Joint Committee on Cancer (AJCC) melanoma staging system: implications for melanoma treatment and care Keung EZ , Gershenwald JE . Expert Review of Anticancer Therapy.2018;18(8). CrossRef
- Tissue biomarkers for prognosis in cutaneous melanoma: a systematic review and meta-analysis Gould Rothberg BE , Bracken MB , Rimm DL . Journal of the National Cancer Institute.2009;101(7). CrossRef
- E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro Tang A, Eller MS , Hara M, Yaar M, Hirohashi S, Gilchrest BA . Journal of Cell Science.1994;107(4). CrossRef
- E-Cadherin Expression in Melanoma Cells Restores Keratinocyte-Mediated Growth Control and Down-Regulates Expression of Invasion-Related Adhesion Receptors Hsu M, Meier FE , Nesbit M, Hsu J, Van Belle P, Elder DE , Herlyn M. The American Journal of Pathology.2000;156(5). CrossRef
- Cyclooxygenase-2 in cancer: A review Hashemi Goradel N, Najafi M, Salehi E, Farhood B, Mortezaee K. Journal of Cellular Physiology.2019;234(5). CrossRef
- High Expression of COX-2 Associated with the Depth of Invasion on Acral Melanoma by Increasing TGF-β1 Gipsyianti N, Aziz A, Hernowo BS , Usman HA . Clinical, Cosmetic and Investigational Dermatology.2021;14. CrossRef
- Prognostic biomarkers of cutaneous melanoma Ding L, Gosh A, Lee DJ , Emri G, Huss WJ , Bogner PN , Paragh G. Photodermatology, Photoimmunology & Photomedicine.2022;38(5). CrossRef
- Functional Roles of Matrix Metalloproteinases and Their Inhibitors in Melanoma Napoli S, Scuderi C, Gattuso G, Bella VD , Candido S, Basile MS , Libra M, Falzone L. Cells.2020;9(5). CrossRef
- Prognostic and Clinicopathological Value of Ki-67 in Melanoma: A Meta-Analysis Liu Q, Peng Z, Shen L, Shen L. Frontiers in Oncology.2021;11. CrossRef
- Expression of E cadherin and Ki 67: Emerging Prognostic Markers in Triple-Negative Breast Cancer Shetty J, Rao C. Indian Journal of Surgical Oncology.2019;10(2). CrossRef
- Vascular endothelial growth factor receptors: Molecular mechanisms of activation and therapeutic potentials Rahimi N. Experimental eye research.2006;83(5). CrossRef
- CD31 and CD34 expression as immunohistochemical markers of endothelial transdifferentiation in human cutaneous melanoma Pisacane A. M., Picciotto F., Risio M.. Cellular Oncology: The Official Journal of the International Society for Cellular Oncology.2007;29(1). CrossRef
- The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets Szklarczyk D, Gable AL , Nastou KC , Lyon D, Kirsch R, Pyysalo S, Doncheva NT , et al . Nucleic Acids Research.2021;49(D1). CrossRef
- Correlation and significance of COX-2, Ki67, VEGF and other immune indexes with the growth of malignant pulmonary nodules Guo H, Xue W, Zhao Q, Zhao H, Hu Z, Zhang X, Duan G. Journal of Cardiothoracic Surgery.2022;17(1). CrossRef
- Loss of E-Cadherin Expression Correlates With Ki-67 in Head and Neck Squamous Cell Carcinoma Dumitru CS , Ceausu AR , Comsa S, Raica M. In Vivo (Athens, Greece).2022;36(3). CrossRef
- Expression of Ki-67 and Estrogen Receptor Beta in Primary Cutaneous Melanoma as a Potential Indicator of Regional Lymph Node Positivity Udovicic-Gagula D, Ahmovic A, Bilalovic N, Doric M. Applied immunohistochemistry & molecular morphology: AIMM.2019;27(1). CrossRef
- Ki-67 expression is superior to mitotic count and novel proliferation markers PHH3, MCM4 and mitosin as a prognostic factor in thick cutaneous melanoma Ladstein RG , Bachmann IM , Straume O, Akslen LA . BMC cancer.2010;10. CrossRef
- Prognostic value of Ki-67 expression in localized cutaneous malignant melanoma Henrique R., Azevedo R., Bento M. J., Domingues J. C., Silva C., Jerónimo C.. Journal of the American Academy of Dermatology.2000;43(6). CrossRef
- The expression of MMP-2 and Ki-67 in head and neck melanoma, and their correlation with clinic-pathologic indices Kamyab-Hesari K, Mohtasham N, Aghazadeh N, Biglarian M, Memar B, Kadeh H. Journal of Cancer Research and Therapeutics.2014;10(3). CrossRef
- Enhanced expression of vascular endothelial growth factor in metastatic melanoma Salven P., Heikkilä P., Joensuu H.. British Journal of Cancer.1997;76(7). CrossRef
- Significance of vascular endothelial growth factor expression in skin melanoma Gajanin V, Krivokuća Z, Kostić K, Gajanin R, Sladojević I. Vojnosanitetski Pregled.2010;67(9). CrossRef
- The role of VEGF in melanoma progression Rajabi P, Neshat A, Mokhtari M, Rajabi MA , Eftekhari M, Tavakoli P. Journal of Research in Medical Sciences: The Official Journal of Isfahan University of Medical Sciences.2012;17(6).
- The role of VEGF-C staining in predicting regional metastasis in melanoma Boone B, Blokx W, De Bacquer D, Lambert J, Ruiter D, Brochez L. Virchows Archiv: An International Journal of Pathology.2008;453(3). CrossRef
- Association of vascular endothelial growth factor expression with patohistological parameters of cutaneous melanoma Gacević M, Jović M, Zolotarevski L, Stanojević I, Novaković M, Miller K, Šuljagić V, Mijušković Ž, Vojvodić D. Vojnosanitetski Pregled.2016;73(5). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2024
Author Details