Ameliorative Potential of Ursolic Acid Isolated from Morina coulteriana Royle Targeting P53 Gene Mutation Induced by Cyclophosphamide
Download
Abstract
Objective: More than 50% of human cancers show mutated TP53 gene. Reactivation or restoration of p53 mutations is viewed as a potential approach for treating cancer. The objective of the present study was to determine whether ursolic acid (UA) could effectively restore the p53 mutations to their wild type or normal state and upregulate its expression.
Methods: The animals were divided into 3 groups, each with 10 mice. Group I was the negative control which received normal saline with 1% dimethyl sulfoxide (DMSO). Group II was the positive control which received cyclophosphamide (CP) (50mg/kg bw) intrapritonealy for 4 days continuously after which the animals were kept on normal saline for the rest 10 days. The treatment group received cyclophosphamide (CP) initially for the first 4 successive days intraperitoneally (50mg/kg bw) and then ursolic acid (UA) (60 mg/kg bw) orally for the next 10 days (post-treatment). Isolated DNA was used in Sanger sequencing of the polymerase chain reaction (PCR) amplified product of the TP53 gene.
Results: Our study demonstrated that UA treatment post CP injection had a restoration efficacy of 89.08%. The molecule, in this investigation, was able to restore the wild type (normal) conformation to the mutations induced by cyclophosphamide in the TP53 gene. 89.08 % mutations converted back to their normal or wild-type forms in the treatment group.
Conclusion: Several investigations have been carried out on this molecule and have indicated UA as a promising chemopreventive agent. The most frequently mutated gene in any type of cancer is the tumor suppressor p53 and UA has shown a potential in functional restoration of the mutated p53 protein. p53 is known to play a vital role in apoptosis and the p53 dependent apoptotic pathway is of prime importance in cancer therapy. Though there has been pioneering work on PRIMA-1 and thiosemicarbazones, identification and development of additional molecules is required.
Introduction
Acting as the major barrier to the malignant transformations and tumor progression, p53 tumor suppressor gene is truly called the “Guardian of the genome”. p53 plays a pivotal role in regulating the cell division and growth, apoptosis and cell cycle blockade, senescence or differentiation in response to varied environmental stresses, thus maintaining the genetic stability. Other roles assigned to p53 include, DNA damage correction, autophagy, proliferation [1-3], inhibition of epithelial to mesenchymal transition and other activities preceding metastasis [4-6].
In recent years it has been studied that the primary target and the frequently altered gene by the environmental mutagens, of most human and animal cancers has been the TP53 gene encoding p53 protein. Mutations in other tumor suppressor genes generally include deletion or truncation leading to cancer. However, the mutations in TP53 allele are predominantly missence mutations leading to the expression of full length p53 mutant protein. Majority of the tumor suppressor genes like RB, APC, BRCA1, get inactivated when mutated, however TP53 when mutated, in addition to the loss of wild type p53 functions, in a dominant negative (DN) manner forms heterotetramers with the unmutated wild type p53 expressed from the other wild type alleles [7, 8]. In short, the mutation in wild type p53 leads to the neomorphic gain of function (GOF) [9] in p53. p53 in this case loses its wild type function and gains a new dominant phenotypic expression which actually contributes to the acquisition of oncogenic properties [10, 11]. Most important addition to it is that the mutation in the TP53 gene could diminish the effectiveness of chemotherapy including agents like doxorubicin, tenozolomide, tamoxifen and gemcitabine [12]. Several studies have explained this occurrence of drug resistance [13]. The inactivation of p53 is the prior requirement for the cancer progression [14-16]. Therefore reactivation of the p53 mutations appears to be an attractive strategy for cancer therapy. Restoration of the wild-type functions should not only re-enable tumor suppressor function but also eliminate the oncomorphic gain of function of the mutant p53. Owing to p53’s critical role in therapy response, in recent years, many promising studies have been proposed where in mutant p53 function has been re-established by certain drug leads [14, 17-21]. One such compound is the APR-246, also known as eprenetapopt or PRIMA-1MET, a derivative of PRIMA 1, which is the most studied of these compounds and has advanced to several clinical studies. Although several such molecules have been identified, additional classes of compounds are highly required.
Cancer has been the leading cause of deaths in the world with 9.6 million deaths recorded in 2018 [22]. Even the conventional therapies for cancer like chemotherapy, radiotherapy, endocrine therapy and targeted therapy have their serious limitations which include severe side effects and dose limiting toxicities [23]. In the past few years, this approach has shifted to the diverse flora breathing in the nature which has shown its potential in treating a huge variety of diseases including cancer from time to time. The chemopreventive agents produced by the plants are the various secondary meatbolites such as polyphenols, isoprenoids, alkaloids, sulphoraphanes and certain vitamins and their precursors [24, 25].
Terpenes are one such class of compounds which have a wide occurrence in the flora on earth [26]. Ursolic acid (UA) is a natural pentacyclic triterpenoid compound possessing diverse pharmacological activities including neuroprotection, anticancer, antimicrobial, hepatoprotection, anti-inflammatory, antioxidative and regulating blood glucose [27-30].
This study involved the study of ameliorative effect of the isolated molecule, ursolic acid from a promising antimutagenic plant Morina coulteriana, on p53 expression against the cyclophosphamide induced mutations.
Materials and Methods
Materials
Cyclophosphamide (CP) was obtained from Sigma Aldrich Co. Oligonucleotide primers were synthesized by Biokart India Pvt. Ltd., Karnataka, India. Ursolic Acid was isolated from the plant Morina coulteriana using chromatographic techniques. Purity of the compound was determined by 13CNMR (Carbon-13 nuclear magnetic resonance) and HPLC (high-performance liquid chromatography). All others chemicals used in the experiment were of analytical grade from Sigma Aldrich.
Experimental design
Thirty, 6-8 week old, Balb/c mice with body weight 25-30 g of both the sexes were used in the experiment. The animals were procured from the Indian Institute of Integrative Medicine (IIIM), Jammu, J & K. The animals were maintained under standard conditions of temperature (22±2º C) and relative humidity (55±10 %) with 12 h light/ dark cycle. The animals were given standard pellet diet and water ad libitum. The whole experiment was carried out according to the guidelines given by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). The experiment was approved by the Institutional Animals Ethics Committee [F (IAEC- Approval) KU/2017/21].
The animals were allocated into 3 groups, each of 10 mice. Group I was the negative control which received normal saline (with 1% DMSO). Group II was the positive control which received cyclophosphamide (50mg/kg bw) intraperitonealy for 4 days continuously after which the animals were kept on normal saline for the rest 10 days. Group III was the treatment group which received CP initially for the first 4 successive days and then UA (60 mg/kg bw) for the next 10 days.
Sampling
24 h post last treatment, 65-100 µl of blood was taken by retro-orbital bleed with a heparinized microcapillary tube from the animals. Blood was immediately stored into a 1.5 ml microfuge tube containing 20 µl of 10 mM EDTA (ethylenediaminetetraacetic acid) and stored at -80º C (for PCR and sequence analysis).
Genomic DNA isolation
DNA was isolated from the blood by retro-orbital bleed using the method adapted from Higuchi (1989) [31].
Evaluation of the quantity and quality of isolated DNA
The amount of DNAisolated was spectrophotometrically (Nanodrop Technologies, Inc., Wilmington, DE, USA) estimated by measuring its optical density (OD) at 260 nm and the purity was assessed as the ratio of absorbance at 260 and 280 nm (A260/A280). The integrity of the isolated DNA was assessed by electrophoresis in 1x Tris-acetate-EDTA buffer (TAE) using 0.8% agarose gel with 2.5µl of ethidium bromide in it. The gel was visualized by a Gel Doc system (Alphaimager TM 2200, Alpha Innotech Corporation) under UV light.
Amplification of the tp53 gene was achieved by polymerase chain reaction (PCR) using a forward primer P53 5´TGCTCACCCTGGCTAAAGTT-3´ and the reverse primer p53 5´AATGTCTCCTGGCTCAGAGG-3´.
DNA sequencing (Sanger Sequencing)
The sequencing was commercially done using the services of Biokart India Pvt. Ltd., Bangaluru. For purification and sequencing, 20 μl of unpurified product samples were given.
Statistical analysis
Average data generated for the treatment group was compared to the respective data of the positive control group. The test results were expressed as mean and standard deviation using Mann-Whitenny U test in Graph- pad prism (10.3.0.)
Results
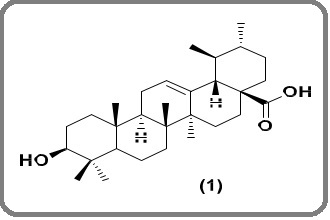
Ursolic acid was isolated from the ethyl acetate fraction of the crude extract of plant Morina coulteriana Royale, collected from Zaznar, Mughal Road, Kashmir (J&K). The plant was identified at the Centre for Biodiversity and Taxonomy and voucher specimen [No. 2634 (KASH)] was preserved in the herbarium. UA was characterized by different spectroscopic techniques and was stored at -20 ºC before being used in the present study. Its structure is shown in Figure 1.
Figure 1. Structure of Ursolic Acid.
As revealed by the Sanger sequence analysis of the amplified PCR products, the negative control group (Group I) which received normal saline (with 1% DMSO) throughout the whole experiment did not show any mutation. The chromatograms from the negative control group show no change in the base sequence hence, no mutations. However the positive control group (Group II) that received the alkylating agent, cyclophosphamide (CP), for 4 days and normal saline for the rest 10 days showed mutations in the TP53. It is because TP53 is considered a major target of CP. The frequency of mutations in the TP53 was majorly (72.41%) transversions and 27.59% tansitions. The major transversions being G→C (28.16%) and G→T (21.83%). However other transversions that occurred were A→C, A→T, C→G. On the other hand, G→A (22.98%) and T→C (4.59%) added to the transition mutations in the gene.
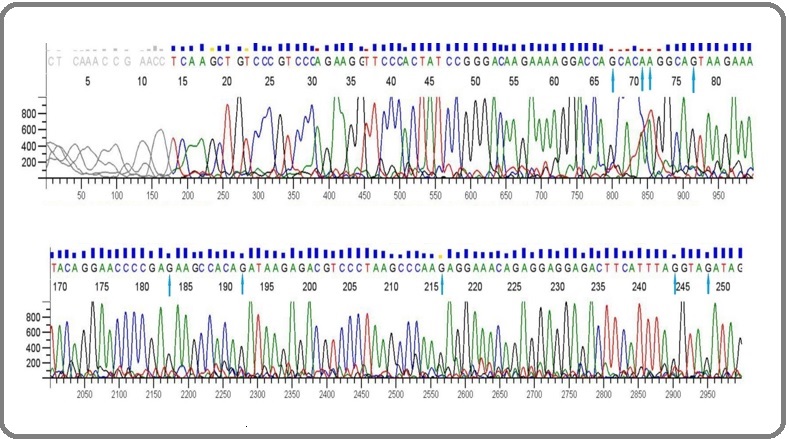
On the other hand, the treatment group that was given CP for the first 4 days and UA for the next 10 days, showed a significant decline in the mutations. In other words, it was able to restore the functionality of the p53 protein by about 89.08 %. Treatment with ursolic acid reversed these mutations to the wild type showing a powerful antimutagenic efficacy. The positions at which the transversions or transitions had occurred showed a reversal to the normal conformation e.g the G→C transversion mutation in the positive control showed a C→G reversal in the treatment group or a G→A transition mutation in the positive control showed A→G reversal in the treatment group. In totality, 89.08 % mutations converted back to their normal or wild-type forms. The difference between the positive control group and the treatment group was recorded to be statistically significant (p<0.0001). The sequencing results for p53 can be viewed in the given chromatograms (Figure 2, 3 and 4).
Figure 2. Electropherogram data showing Sanger sequencing of p53 gene in the negative control group. The arrows show the sites at which mutations occur in the positive control group when CP is administered.
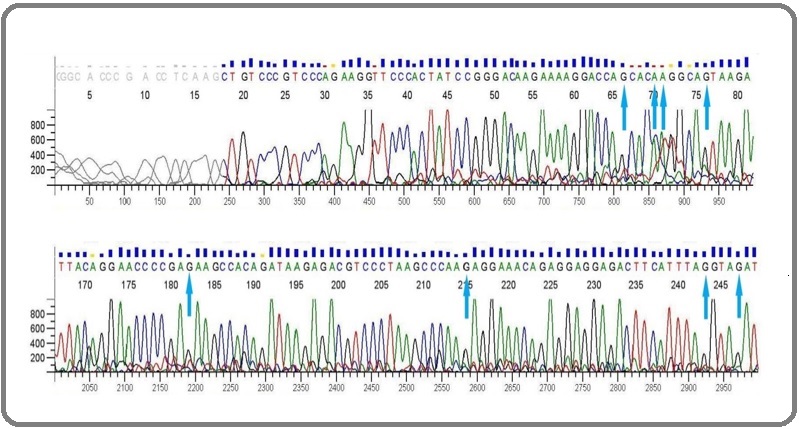
Figure 3. Electropherogram data showing Sanger sequencing of p53 gene in the positive control group. The arrows show the mutation sites (G→C, A→C, A→T, G→T, G→A) .
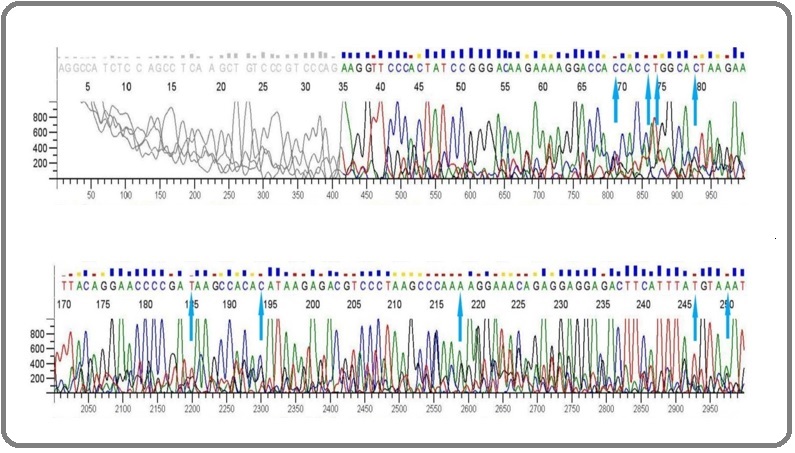
Figure 4. Electropherogram data showing Sanger sequencing of p53 gene in the treatment group. The arrows show the sites where the mutations have reverted (C →G, C→A, T→A, T→G, A→G).
Discussion
Research in the last 40 years, has established that the p53 tumor suppressor plays a significant role in preventing the growth of tumors and neoplastic transformation by acting as an exceptionally sensitive stress sensor and coordinating a complex network of various effector pathways and processes that safeguard genome stability and cellular homeostasis. Also, in human malignancies, missense mutations in the TP53 gene are incredibly common. These mutations result in mutant p53 proteins, some of which exhibit trans-dominant suppression over their wild type counterparts and lose their tumor suppressor properties.
Mutations in p53 could further diminish the effectiveness of doxorubicin, tamoxifen and other chemotherapeutic agents [12]. p53 has proven to be a promising target in the quest for novel anticancer strategies. p53 is considered as a major target of the chemotherapeutic drug cyclophosphamide (CP) which is an alkylating agent [32] and has been used in the present study. Numerous studies have demonstrated that the bioactive components from plants could have effectiveness in the battle against cancer. Curcumin, for example, has been studied to inhibit the mutations induced by cyclophosphamide in exon 7 of the TP53. Also, chlorophyllin has been found to have a powerful protective efficacy of 80% against CP induced mutations in the TP53 exon 7 [33]. UA, one such phytochemical has been studied to play a promising role in chemoprevention. Various investigations till now support this statement. Present study shows an immense chemoprotective role of UA against CP. Inspite of having low solubility and poor bioavailability, UA possess a vast array of pharmacological and toxicological activities. Future experiments are needed to further work on the mechanics involved in the restoration of wild-type- like properties to the p53 hot spot mutations. From all its reasonable to conclude that UA has shown a clear antimutagenic potential against cyclophosphamide in reverting the mutations in p53 gene.
Acknowledgments
The authors are thankful to the Director, Centre of Research for Development, University of Kashmir for showing interest in this research work and permitting to carry out the research. The authors are also thankful to the Centre for Biodiversity and Taxonomy, University of Kashmir for the identification of the plant specimen.
Funding statement
This study did not receive any funding and the cost of research was met with by the authors.
Conflict of Interest
Authors declare no conflict of interest.
Ethical declaration
The study was approved by the Institutional Animals Ethics Committee [F (IAEC-Approval) KU/2017/21].
Author contribution
All authors contributed equally.
Data availability
The article contains all the essential data. Any other data or supporting documents etc. are available on request.
References
- Cancer. p53, guardian of the genome Lane D. P.. Nature.1992;358(6381). CrossRef
- p53, the cellular gatekeeper for growth and division Levine A. J.. Cell.1997;88(3). CrossRef
- Twenty years of p53 research: structural and functional aspects of the p53 protein May P., May E.. Oncogene.1999;18(53). CrossRef
- The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression Chen J. Cold Spring Harbor Perspectives in Medicine.2016;6(3). CrossRef
- Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe Klymkowsky MW , Savagner P. The American Journal of Pathology.2009;174(5). CrossRef
- The molecular mechanism of action of methylene quinuclidinone and its effects on the structure of p53 mutants Omar SI , Tuszynski J. Oncotarget.2018;9(98). CrossRef
- p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM Song H, Hollstein M, Xu Y. Nature Cell Biology.2007;9(5). CrossRef
- Context is everything: extrinsic signalling and gain-of-function p53 mutants Amelio I, Melino G. Cell Death Discovery.2020;6. CrossRef
- Mutant p53 in cancer: new functions and therapeutic opportunities Muller PAJ , Vousden KH . Cancer Cell.2014;25(3). CrossRef
- When mutants gain new powers: news from the mutant p53 field Brosh R, Rotter V. Nature Reviews. Cancer.2009;9(10). CrossRef
- Mutant p53 gain-of-function in cancer Oren M, Rotter V. Cold Spring Harbor Perspectives in Biology.2010;2(2). CrossRef
- The role of p53 in cancer drug resistance and targeted chemotherapy Hientz K, Mohr A, Bhakta-Guha D, Efferth T. Oncotarget.2017;8(5). CrossRef
- Towards the overcoming of anticancer drug resistance mediated by p53 mutations Cao X, Hou J, An Q, Assaraf YG , Wang X. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy.2020;49. CrossRef
- Restoration of p53 function: a new therapeutic strategy to induce tumor regression? Beraza N, Trautwein C. Hepatology (Baltimore, Md.).2007;45(6). CrossRef
- Modeling the therapeutic efficacy of p53 restoration in tumors Martins CP , Brown-Swigart L, Evan GI . Cell.2006;127(7). CrossRef
- Restoration of p53 function leads to tumor regression in vivo Ventura A, Kirsch DG , McLaughlin ME , Tuveson DA , Grimm J, Lintault L, et al . ResearchGate.2024. CrossRef
- Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound Bykov VJN , Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG , Selivanova G. Nature Medicine.2002;8(3). CrossRef
- Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs Bykov VJN , Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG . The Journal of Biological Chemistry.2005;280(34). CrossRef
- Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53 Wassman CD , Baronio R, Demir Ö, Wallentine BD , Chen C, Hall LV , Salehi F, et al . Nature Communications.2013;4. CrossRef
- Allele-specific p53 mutant reactivation Yu X, Vazquez A, Levine AJ , Carpizo DR . Cancer Cell.2012;21(5). CrossRef
- Mutant p53 targeting by the low molecular weight compound STIMA-1 Zache N, Lambert JMR , Rökaeus N, Shen J, Hainaut P, Bergman J, Wiman K , Bykov VJN . Molecular Oncology.2008;2(1). CrossRef
- Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray F, Ferlay J, Soerjomataram I, Siegel RL , Torre LA , Jemal A. CA: a cancer journal for clinicians.2018;68(6). CrossRef
- Trametes robiniophila Murr in the treatment of breast cancer Li C, Wang X, Chen T, Wang W, Yang Q. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie.2020;128. CrossRef
- Dietary antioxidant vitamin C influences the evolutionary path of insecticide resistance in Drosophila melanogaster Huang J, Sun W, Seong KM , Mittapalli O, Ojo J, Coates B, Paige KN , Clark JM , Pittendrigh BR . Pesticide Biochemistry and Physiology.2020;168. CrossRef
- Plasma protein binding of dietary polyphenols to human serum albumin: A high performance affinity chromatography approach Cao H, Liu X, Ulrih NP , Sengupta PK , Xiao J. Food Chemistry.2019;270. CrossRef
- Signalling pathways of the TNF superfamily: a double-edged sword Aggarwal BB . Nature Reviews. Immunology.2003;3(9). CrossRef
- Ursolic acid: A systematic review of its pharmacology, toxicity and rethink on its pharmacokinetics based on PK-PD model Sun Q, He M, Zhang M, Zeng S, Chen L, Zhou L, Xu H. Fitoterapia.2020;147. CrossRef
- Interference of ursolic acid treatment with glioma growth: An in vitro and in vivo study Bergamin LS , Figueiró F, Dietrich F, Manica FDM , Filippi-Chiela EC , Mendes FB , Jandrey EHF , Lopes DV , Oliveira FH , Nascimento IC , Ulrich H, Battastini AMO . European Journal of Pharmacology.2017;811. CrossRef
- Recent developments on the extraction and application of ursolic acid. A review López-Hortas L, Pérez-Larrán P, González-Muñoz MJ , Falqué E, Domínguez H. Food Research International (Ottawa, Ont.).2018;103. CrossRef
- Ursolic acid enhances the cellular immune system and pancreatic beta-cell function in streptozotocin-induced diabetic mice fed a high-fat diet Jang S, Yee S, Choi J, Choi M, Do G, Jeon S, Yeo J, Kim M, Seo K, Lee M. International Immunopharmacology.2009;9(1). CrossRef
- Rapid, efficient DNA extraction for PCR from cells or blood Higuchi R. Perkin Elmer/Cetus Newsletter.1989;1.
- Post‐translational modifications and activation of p53 by genotoxic stresses Appella E, Anderson CW . Eur J Biochem.2001;268(10):2764-72. CrossRef
- Antimutagenic efficacy of some natural compounds on cyclophosphamide-induced p53 alterations Gouda EM , Elbehairy AM , Ghoneim MA . Zeitschrift Fur Naturforschung. C, Journal of Biosciences.2008;63(11-12). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2024
Author Details