Using High Resolution Melting Method for Detecting Nucleotide Polymorphisms of STK11 Gene Germline in Patients with Digestive System Cancers
Download
Abstract
Background and Objective: High resolution melting (HRM) analysis is a technique to measure decrease of fluoresce, caused by discharge of the dye, throughout DNA thermal melting gradient process. STK11 gene encodes one of cellular serine-threonine kinase proteins that regulates cellular polarity. It also acts as tumor suppressor protein. Mutations of germline in the gene is coincident with Peutz-Jeghers syndrome and potential of developing a variety of neoplasia.
Methodology: In an experimental study, genome DNA of 56 patients with digestive system cancer was extracted. Afterward, nucleotide changes over the gene were examined using Real-time PCR and High resolution melting (HRM).
Findings: Nucleotide screening using HRM technique revealed two types of SNP in introns 6 & 7 in 10 patients. Four patients showed homozygous C/T nucleotide changes [cluster id/dsSNP/rs9282860] in intron 6 and six patients showed heterozygous C/G nucleotide changes [cluster id/dsSNP/rs2075607] in intron 7. Comparison of HRM results with sequencing results indicated 100% conformity.
Conclusion: Although no mutation was observed in exon section of the gene, primary screening of STK11 gene to diagnose unknown nucleotide changes of germline and somatic in patients with neoplasia using HRM was feasible, easy, and cost effective.
Introduction
Following PCR proliferation, high resolution melting (HRM) is a new and homogenous method, which is performed in a closed tube. This method is feasible in PCR products for analyzing genetic changes (SNPs, mutation, and methylation). HRM differentiates nucleic acid samples based on sequence, length, and GC volume. It is featured with proliferation of the gene in 80-250bp pieces in a reaction in which florescent dyes are bound to two-stranded DNA. Following PCR, and as HRM process goes on, the double-stranded DNA melts and the dye fluoresces at a lower level. The process is plotted in a melt curve with high resolution that is highly variable for sequences of DNA so that it is possible to distinguish amplicons that are different even only in one unbound pair. HRM analysis can be used for detecting small genetic changes such as insertion, deletion, and substitution of single nucleotide polymorphism (SNP) [1].
Comparing with other SNP diagnosis technologies such as sequencing, denaturing higher pressure liquid chromatography (DHPLC), and denaturing gradient gel electrophoresis (DGGE), HRM is easier to implement, more cost/time effective, and needs less volume of reaction agents. Moreover, HRM is a closed-tube process, which minimizes risk of contamination of PCR products. Since HRM analysis is a non-invasive method, it can directly sequence products of HRM reaction with no need for additional proliferation or purification [2, 3].
STK11 or LKB1 gene was discovered in 1998 through linkage analysis in Peutz-Jeghers syndrome (PJS) as suppressor of tumors [4]. PJS is a rare autosomal disorder with decreased prevalence that was first diagnosed in a Danish family by Peutz in 1921 and later was explained by Jeghers et al. in 1949. In addition, to mucus and cutaneous pigments and gastric-intestinal polypose hamartomateuse there is a risk of development of benign and malignant tumors in digestive limbs and other areas like chest, intestine, testicle, cervix, lungs, pancreases, and skin. Deactivation of STK11 gene in PJS patients reaches to 91%, out of which 78% is due to point mutation and 13% is due to deletion [4, 5].
STK11 gene demonstrates serine/threonine kinase activity in 19p13.3 chromosome. It expanses in 23kbase region with 9 encoder exons and one non-encoding exon. mRNA of the gene is 2.4kbase in length and transcribed in telomere – centromere. This gene is translated into a protein with 433 amino acid with molecular weight of 48kd. The protein is featured with peptide signal at the end of amino acid 338-43 and a kinase domain (amino acid 49-309) [4, 6].
STK11 gene is expressed in all body tissues both in fetus and adults and liver, pancreases, testicle, and skeletal muscles in particular. The product of STK11 is the first upstream kinase for adenine monophosphate-activated protein kinase (AMPK) that is activated allosterically through attaching to a pseudokinase (STRAD) and an adaptor protein (MO25). Hetromic set of STK11-STRAD-MO25, as an active biological unit is capable of phosphorylation and activation of AMPK and 12 other kinases at least [4, 7, 8]. In fact, AMPK is the essential part of the cellular metabolism that is needed to preserve cellular hemostasis. AMPK activation by STK11 leads to suppression of growth and proliferation when nutrient level decreases. Moreover, activation of AMPK-like kinases by STK11 plays a key role in preserving cellular polarity, which results in suppression of tumor cells.
In short, STK11 gene is a key element in cellular polarity, metabolism, p53 depended apoptosis, inhibition of cellular cycle in G1 and cell proliferation. Thereby, in addition to PJS, any disorder in the gene leads to prevalent disorders like diabetes and cancer [9].
Materials and Methods
To examine mutation of STK11 gene germline, complete blood samples of 56 patients (different ethnic groups) with cancer of digestive system and age range of 45-80 were procured, with observance of moral codes, from Imam Khomeni and Javaheri Hospitals in Tehran and BuAli Hospital in Hamedan. The samples were stored at -20ºC and then used for extracting genome DNA.
Genome DNA was extracted from 200µl complete blood using High pure PCR Template Preparation kit (Roche) according to manufacturer’s recommended process.
Proliferation of 10 exons of STK11 gene was done using the primers that have been studied for this gene [10]. Twenty pairs of primers were synthesized by TAG Copenhagen. Primer sequence and size of each proliferated piece in PCR reaction are listed in Tabl 1.
| Gene | Exon | Sence primer | Antisense primer | Size amplicon (bp) |
| STK11 | 1 | CGAGCTGATGTCGGTGGGT | ACAGCGTCTCCGAGTCCAG | 193 |
| STK11 | 2 | ATCATCCTGACGTTGGGTCG | GGAGACGGGAAGAGGAGCAG | 248 |
| STK11 | 3 | CCCTCCAGAGCCCCTTTT | CACGCTGTCCAGCATTTCC | 161 |
| STK11 | 4-5 | CGGTGGCACCCTCAAAAT | GACCCCAGCCGACCAGAT | 267 |
| STK11 | 6 | TGGGTCCAGAGGACACTCCC | CCCTTCCCGATGTTCTCAAA | 291 |
| STK11 | 7 | CCGGCTTCTGGGCGTTTGC | TCTAGCGCCCGCTCAACC | 174 |
| STK11 | 8 | CTGCTTCTGGGCGTTTGC | ACCGTGAAGTCCTGAGTGTAGAT | 220 |
| STK11 | 9 | CTTGCCGTCTCCCTCCCA | TCCGCCCTGGATTTGGTG | 151 |
| STK11 | 10 | GAGTCCGGTAGCCCCATGA | TGGTCGGCACAGAAGCATG | 237 |
To optimize PCR and HRM reactions, PCR was performed for all 10 pairs of primers in identical condition explained as follows. Total volume of reactor was 25µl in volume containing genome DNA (100-200 ngr), primers (1µmol each), dNTPs (200µmol), MgCl2 (1.5mmol), DMSO (10%), Hot Start Taq DNA polymerase (Promega) (2.5units), and SYTO-9 (Invitrogen, 2µmol). Heating plan included primary denaturation (5min, 95ºC) followed by 40 thermal cycles (25s-95ºC & 20S 50ºC) along with adsorbing the dye at green channel (30S, 72ºC) and HRM (75ºC – 93ºC) with temperature shift of 0.1ºC/s.
All the samples and exons that showed nucleotide differences based on HRM results and the cases that showed no nucleotide difference were sequenced by Forward primer and Reserve primer supplied from Macrogen (Korea) and nucleotide changes by DNA Sequencing.Nucleotide change C/T [cluster id/dsSNP/rs9282860] in intron 6 was homozygous and part of class 1 - according to SNP classification [2].Six patients had heterozygous nucleotide changes C/G [cluster id/dsSNP/rs2075607] in intron, which was in class 3. (Fig. 1-4) illustrate HRM image and SNP sequencing. Sequencing results based on HRM technique was completely identical to that of other samples without nucleotide change.
Figure 1 :Sequencing Nucleotide Changes In Exon/Intron 6 Border
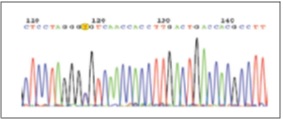
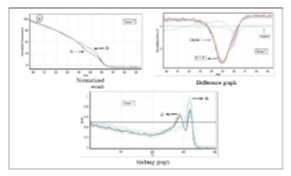
Figure 2 : HRM Results And The Generated Curves (Normalized/Difference/Melting Graphs) Of STK11 Gene On Exon/Intron 6 Border In Rotor-Gene 6000

Figure 3 :: Sequencing Nucleotide Changes In Exon/Intron 7 Border
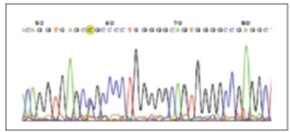
Figure 4 :HRM Results And The Generated Curves (Normalized/Difference/Melting Graphs) Of STK11 Gene On Exon/Intron 7 Border In Rotor-Gene 6000
Discussion
High resolution DNA melting is powerful and new method for screening diversity of nucleotides. Although sensitivity and specificity of the method depends on variety of factors such as length of PCR product, type of dye, density of Mg2+, it still outperforms other techniques for detecting unknown mutation. Advantages of HRM are ease of implementation, accuracy, and time efficiency so that there is no need for Post-PCR processes and the risk of secondary contamination is eliminated [1, 11]. To examine sequence of STK11 gene in exons and exon/intron border, HRM technique was used. The participants were anonymous and all diagnosed with digestive system cancer. Among 56 participants, only 10 (four with intron 6 and six with intron 7) demonstrated two types of nucleotide changes and no correlation between these two have been reported thus far. HRM results were consistent with PCR sequencing.
A key feature of this technique is the possibility of distinguishing homozygous and heterozygous cases using melting graph. As illustrated in (Fig. 2-4), heterozygoutes are distinguished by two peaks and homozygoutes by one peak. Additionally and in presence of DMSO (10%), the reaction creates more sensitivity in diagnosing single nucleotide differences in a thermal change step of 0.2ºC/s. At any rate, mutation of germline in STK11 encoding regions was not examined, which does not mean there was no mutation in STK11 gene. Given the complicated and variant role of STK11 gene in function of cells, future studies can examine the gene germline in other patients including diabetics.
Acknowledgements
This research work is part of thesis work of Seyed Jamal HosseiniAli Nazemi. This research plan was financially supported by Biology Association of The Islamic Azad University-Tonekabon. The authors wish to thank Dr. Arash Jenabian, Dr. Mahmoud Abbasi, Mohammad Yaghoub Taleghani, and Rouzbeh Tabib for their help in collecting the samples.
References
[1]. Taylor CF. Mutation scanning using high resolution melting. Biochem Soc Transact 2009; 37(pt 2): 433-37.
[2]. Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50(7):1156-64.
[3]. Kristensen LS, Dobrovic A. Direct genotyping of single nucleotide polymorphisms in methyl metabolism genes using probe-free high-resolution melting analysis. Cancer Epidemiol Biomarkers Prev 2008; 17(5): 1240–47.
[4]. Fan D, Ma C, Zhang H. The molecular mechanisms that underlie the tumor suppressor function of LKB1. Acta Biochim Biophys Sin 2009; 41(?): 97–107.
[5]. Schumacher V, Vogel T, Leube B, Driemel C, Goecke T, Moslein G, et al. STK11 genotyping and cancer risk in Peutz-Jeghers syndrome. J Med Genet. 2005;42(5):428-35.
[6]. Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, Shekher MC, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154(6):1835-40.
[7]. Wang ZJ, Churchman M, Avizienyte E, McKeown C, Davies S, Evans DG, et al. Germline mutations of the LKB1 (STK11) gene in Peutz-Jeghers patients. J Med Genet. 1999;36(5):365-8.
[8]. Yoo JH, Choi YJ, Kang JG, Sun YK, Ki CS, Lee KA. A novol de novo mutation in the serine-threonine kinase STK11 gene in a Korean patient with Peutz-Jeghers syndrome. BMC Med Genet.2008; 9: 1-5.
[9]. Karuman P, Gozani O, Odze RD, Zhou XC, Zhu H. The Peutz-Jegher gene product LKB1 is a mediator of p53- dependent cell death. Mol Cell.2001; 7(6): 1307-19.
[10]. Meur NL, Martin C, Veber PS, Joly GR, Oise F, Lemoine M, et al. Complete germline deletion of the STK11 gene in a family with Peutz–Jeghers syndrome. Eur J Human Genet .2004; 12(5): 415–18.
[11]. Laurie AD, George PM. Evaluation of high-resolution melting analysis for screening the LDL receptor gene. Clin Biochem. 2009;42(6):528-35.
[12]. Movafagh A, Hajifathali A, Isfahani F, et al. Geographic heterogeneity of cytogenetic characteristics of acute myeloid leukemia in the early detection. Iran J Cancer Prev. 2012;2: 85-9.
[13]. Movafagh A, Mirfakhraei R, Mousavi-Jarrahi A. Frequent incidence of double minute chromosomes in cancers, with special up-to-date reference to leukemia. Asian Pac J Cancer Prev. 2011;12(12):3453-6.
[14]. Movafagh A, Hajifathali A, Zamani M. Secondary chromosomal abnormalities of de novo acute myeloid leukemia--a first report from the Middle East. Asian Pac J Cancer Prev. 2011;12(11):2991-4.
[15]. Fardmanesh H, Shekari M, Movafagh A, Alizadeh Shargh S, Poursadegh Zonouzi AA, Shakerizadeh S, et al. Upregulation of the double-stranded RNA binding protein DGCR8 in invasive ductal breast carcinoma. Gene. 2016;581(2):146-51.
[16]. Shargh SA, Sakizli M, Khalaj V, Movafagh A, Yazdi H, Hagigatjou E, et al. Downregulation of E-cadherin expression in breast cancer by promoter hypermethylation and its relation with progression and prognosis of tumor. Med Oncol. 2014;31(11):250.
License
Copyright
© ,
Author Details