Dual Drug Repurposing in Cervical Cancer: The Synergistic Cytotoxic Effect of Dapagliflozin-Etoricoxib and Its Predicted Modulation of PI3K/Akt/mTOR Signaling via Molecular Docking
Download
Abstract
Objective: This study assesses the effectiveness of combining Dapagliflozin and etoricoxib in inhibiting cancer cell growth and investigates its effects on the mutant PI3K/Akt/mTOR signaling pathway to understand the underlying mechanisms.
Methods: After incubation periods of 24 and 72 hours, HeLa cells and normal human fibroblasts (NHF) were utilized to assess the anticancer efficacy and safety profile of Dapagliflozin, Etoricoxib, their combination, and 5-fluorouracil (5FU). The tested concentrations ranged from 0.1 to 1000 µg/ml. To determine potential synergy and selectivity, the combination index (CI) and the selective toxicity index (SI) were estimated. Additionally, molecular docking simulations were performed to evaluate the binding affinities of Dapagliflozin and Etoricoxib to mutant proteins within the PI3K/Akt/mTOR signaling pathway.
Results: The MTT assays showed that a combination of Dapagliflozin and etoricoxib has significant anticancer activity. The mixture effectively inhibits the growth of cervical cancer cells, achieving results similar to 5-fluorouracil (5FU) and outperforming Dapagliflozin or etoricoxib alone. Additionally, the cytotoxic effects of the mixture on normal human fibroblast (NHF) cells were much lower than those seen with 5FU, indicating decreased toxicity. The combined use of Dapagliflozin and etoricoxib exhibited synergistic cytotoxic effects, as indicated by the combination index (CI) score. This drug pair also showed selectivity in targeting cancer cells, as reflected by the selectivity index (SI). The molecular docking results showed that Dapagliflozin and Etoricoxib have affinities for interacting with the mutant PI3K/Akt/mTOR signaling protein. Docking scores for Dapagliflozin binding to these proteins were -8, -6.7, and -7.3 kcal/mol, while those for Etoricoxib were -8, -6.6, and -6.8 kcal/mol, respectively.
Conclusion: The findings, supported by established pharmacokinetic and safety data, suggest that combining Dapagliflozin and Etoricoxib may provide a safer and more effective treatment option for cervical conditions, with a possible mechanism involving the PI3K/Akt/mTOR pathway, as predicted by molecular docking.
1. Introduction
Cervical cancer represents a significant global public health challenge, particularly in low- and middle-income countries (LMICs), where it is the fourth most prevalent malignancy [1]. Infections caused by high-risk human papillomavirus (HPV) types, especially HPV-16 and HPV-18, account for approximately 70% of cases [2, 3]. Even though there are good programs for screening and HPV vaccines, people in low- and middle-income countries have less access, leading to higher illness and death rates compared to richer countries [3, 4]. Early adoption of Pap smears and HPV testing has helped reduce cervical cancer in regions with good healthcare. However, universal access to prevention and treatment remains a global challenge [3, 5-9]. Chemoradiotherapy constitutes the primary therapeutic approach for cervical carcinoma. The chemotherapy component, typically employing cisplatin-based regimens, presents considerable challenges such as systemic toxicity, the development of drug resistance, and detrimental effects on adjacent healthy tissues [10, 11]. Side effects, including nephrotoxicity, myelosuppression, and gastrointestinal distress, frequently diminish patients’ quality of life and may restrict continuing treatment [12].
The adverse effects associated with chemotherapy underscore the critical necessity for safer therapeutic alternatives. Extensive research has been conducted on the potential repurposing of existing pharmacological agents, initially developed for various therapeutic applications, to identify efficacious treatment options for cervical ailments [13-22]. Several studies were conducted in conjunction with this concept, one of which demonstrated that combining esomeprazole with amygdalin exhibits a potent synergistic cytotoxic effect against HeLa cervical cancer cells in vitro [13]. Meanwhile, another study demonstrated that combining vinblastine with laetrile synergistically inhibits the proliferation of HeLa cervical cancer cells in vitro [22]. Furthermore, recent research has demonstrated that the synergistic combination of esomeprazole and ciprofloxacin effectively inhibits the proliferation of HeLa cells by targeting the heat shock protein 70 (Hsp70) [23]. Aligned with this concept, Dapagliflozin and Etoricoxib are examples of drugs with potential anticancer properties. Their selection was based on comprehensive pharmacokinetic analyses and safety assessments.
Moreover, numerous prior studies have demonstrated their anticancer activity.
Recent findings suggest that Dapagliflozin may offer benefits beyond managing diabetes. In vivo and in vitro studies have shown that dapagliflozin can reduce cancer cell growth and trigger apoptosis, especially in models of breast and prostate cancer [24]. The observed effects are due to the drug’s ability to disrupt glucose metabolism in cancer cells, thereby impairing their energy supply and inhibiting growth [25]. Furthermore, SGLT2 inhibitors appear to influence key cell signaling pathways, including the PI3K/AKT/mTOR pathway, which is essential for tumor survival [26]. These mechanisms indicate that dapagliflozin might be used together with standard cancer treatments to improve their effectiveness and lessen metabolic side effects. However, different results seen in certain cancers, such as liver cancer, suggest that the drug’s effect may vary [26].
Dapagliflozin (C₂₁H₂₅ClO₆), a selective SGLT2 inhibitor, features a thiazolidinedione-derived core structure with a dichlorophenyl group at the C-5 position and a β-D-glucopyranosyl moiety at the N-terminus. This unique architecture provides glucose-lowering effects through renal glucose excretion and may also exhibit potential off-target anticancer activity by modulating metabolic pathways. The compound’s lipophilic aromatic ring system and hydrophilic sugar moiety contribute to its optimal pharmacokinetic profile and tissue distribution [27, 28]. Recent studies suggest that structural similarities between dapagliflozin’s thiazolidinedione component and known PPARγ modulators may explain its secondary pharmacological effects [29] .
Conversely, etoricoxib exhibits antiproliferative and pro-apoptotic effects across various cancer cell lines, including those of colorectal, breast, and lung cancers [30]. The observed reduction in tumor growth may be due to etoricoxib’s ability to inhibit the synthesis of prostaglandin E2 (PGE2), a key factor in the development of inflammation-related cancers. This inhibition may play a crucial role in mitigating the inflammatory processes that contribute to tumor progression [31]. Etoricoxib, by inhibiting COX-2-related PGE2 production, disrupts key oncogenic pathways including NF-κB and Wnt/β-catenin signaling. These pathways are well-known for promoting cell survival, angiogenesis, and metastasis, highlighting the potential of etoricoxib in targeting tumor progression [32]. Additionally, another study finding indicates that etoricoxib enhances the effectiveness of standard chemotherapy drugs, likely by reducing drug resistance related to the microenvironments of inflammatory tumors [33]. However, the effectiveness of various anticancer agents seems to vary depending on the molecular features of the tumor, especially COX-2 expression levels. cancers with high COX-2 expression, like colorectal cancer, often respond better to certain treatments than those with low COX-2 activity. This difference underscores the importance of personalized medicine approaches that take into account tumor biology when developing treatment strategies [34].
Etoricoxib (C₁₈H₁₅ClN₂O₂S) is a selective COX-2 inhibitor characterized by a methylsulfonylphenyl moiety attached to a pyridine backbone with a chlorophenyl substituent at the 5-position [35]. This unique structure confers high COX-2 selectivity (IC₅₀ = 1.1 μM) while minimizing COX-1 inhibition (IC₅₀ > 100 μM) [36]. The planar, lipophilic nature of its dipyridine core facilitates optimal binding to the COX-2 active site, while the para-methylsulfonyl group enhances anti-inflammatory potency [35].
The PI3K/Akt/mTOR signaling pathway plays a crucial role in controlling cell growth, proliferation, and survival, and it is often dysregulated in many cases of cancer [37]. Activation of this cascade begins with PI3K-mediated generation of PIP3, which recruits and activates Akt, subsequently phosphorylating mTOR to promote protein synthesis and metabolic reprogramming [38]. In cancer, hyperactivation occurs through genetic alterations, such as PIK3CA mutations, PTEN loss, or amplification of upstream receptor tyrosine kinases, driving uncontrolled proliferation and therapeutic resistance [39]. The pathway’s oncogenic effects extend to angiogenesis, metastasis, and immune evasion, making it an attractive therapeutic target [40]. However, clinical success with PI3K/Akt/mTOR inhibitors has been limited by compensatory feedback mechanisms and toxicity, prompting investigation into combination therapies and isoform-specific agents [41]. Recent advances highlight the potential of biomarker-driven approaches to improve therapeutic outcomes for patients with pathway-activated tumors [42].
While many studies examine the anticancer effects of Dapagliflozin and etoricoxib individually, they have not explored the potential of their combination or its impact on the PI3k/Akt/mTOR pathway. This research aims to fill this gap by evaluating the anticancer properties of the Dapagliflozin-etoricoxib mixture and investigating its effect on key molecular mechanisms, especially its ability to target the PI3k/Akt/mTOR pathway.
2. Materials and Methods
2-1- Study medication
The medications used in the study (Dapagliflozin and etoricoxib) were sourced as raw materials from the Samarra Pharmaceutical Factory in Iraq. Both drugs and their mixture underwent serial dilution using MEM media, resulting in concentrations ranging from 0.1 to 1000 µg/ ml. The concentrations of Dapagliflozin and etoricoxib in the mixture varied from 0.05 to 50 µg/mL for each drug, resulting in a final concentration between 0.1 and 1000 µg/mL.
2-2- Cytotoxicity assay
A cytotoxicity assessment was conducted using the human cervical cancer cell line (HeLa) to investigate the anticancer properties of Dapagliflozin, etoricoxib, 5FU, and the Dapagliflozin-etoricoxib mixture. Additionally, the cytotoxic effects of the mixture on the NHF cell line, regarded as a “normal healthy cell line,” were examined to assess its safety and identify any potential drug interactions that might adversely affect normal cells. The cytotoxic and safety profiles of Dapagliflozin, etoricoxib, 5FU, and the mixture were evaluated by measuring the viability of cancerous and healthy normal cells at concentrations ranging from 0.1 to 1000 µg/ml.
All cancer cell lines utilized in this study were procured from the tissue culture unit at ICCMGR.
2-2-1- Cell Lines
Cancerous and healthy cell lines were utilized, including the HeLa cell line derived from human cervical cancer tissue [43, 44]. The NHF cell line is derived from normal human adipose tissue [45].
2-2-2- Tissue culture conditions
MEM media (US Biological, USA) was used for cell line growth. It was supplemented with 10% (v/v) fetal bovine serum (FBS) (Capricorn-Scientific, Germany), 100 IU of penicillin, and 100 µg of streptomycin to prevent bacterial contamination. The cells were maintained in a humid environment at 37 °C during exponential growth. [46].
2-2-3- cytotoxicity study
The MTT assay is a colorimetric test that leverages the ability of living cells to transform MTT stain into purple formazan crystals, a reaction facilitated by mitochondrial dehydrogenase enzymes. Typically, cells are cultured in a 96-well plate and exposed to varying concentrations of test substances. Following an incubation period, MTT is added to each well and the plate is incubated again. Viable cells convert MTT into formazan, which can be solubilized, and its concentration is assessed by measuring the absorbance at a specific wavelength using a spectrophotometer.
The number of viable cells affects the extent of formazan production. A decrease in formazan production following the application of the investigated medications suggests cytotoxicity, subsequently influencing absorbance levels. The dose-response curve is utilized to determine the half-maximal inhibitory concentration (IC50), which denotes the concentration of medications that reduces cell viability by 50%. This calculation is performed using GraphPad Prism (version 9.5.0 (750)) [19, 47].
Cells were seeded in a 96-well microplate at a density of 10,000 cells per well and incubated at 37 °C for 24 hours until reaching confluence. The MTT cytotoxicity assay included six wells for each concentration of Dapagliflozin, etoricoxib, 5FU, and (Dapagliflozin–etoricoxib) mixture. Cells were treated with concentrations of 0.1, 1, 10, 100, and 1000 µg/ml, while several untreated wells served as a negative control. After 24 and 72 hours of treatment, 28 µL of MTT dye solution (2 mg/ml) was added to each well and incubated for three hours. Subsequently, 100 μL of DMSO was added to each well and incubated for 15 minutes. Optical density readings were obtained at 570 nm using a microplate reader. The following equation is used to calculate the growth percentage inhibition [48].
Growth inhibition %= (optical density of control wells-optical density of treated wells)/(optical density of control wells)*100%
2-3- Selective toxicity index
The selective toxicity index score was computed to assess the selective toxicity of the (Dapagliflozin– etoricoxib) mixture and 5FU on cancer cells following 24 and 72 hours of incubation. After establishing the IC50 levels for the mixture and 5FU, the selective cytotoxicity index was calculated using a mathematical formula based on cellular growth curves for HeLa and NHF cell lines [49].
Selective toxicity Index (SI)=(IC50 ofnormal cell lines)/ (IC50 ofcancer cell lines)
An SI score above 1.0 signifies a drug’s enhanced ability to target tumor cells compared to its toxicity towards normal cells.
2-4- Molecular docking
The chemical structures of Dapagliflozin and etoricoxib were created and optimized using the ChemDraw application (Cambridge Soft, USA) and Chem3D. The chemical docking of these compounds with mutant signal proteins of the PI3K-Akt-mTOR pathway was performed. The molecular structures of each mutant protein, sourced from the Protein Data Bank under the codes (PDB:6oac), (PDB:6s9k), and (PDB:4jsv), pertain to the mutant PI3K-Akt-mTOR, respectively. These mutant structures were chosen for docking studies due to their representation of constitutively active, disease-relevant conformational states, which serve as primary targets for therapeutic inhibition in oncology.
Mutant signal protein structures were optimized and refined using AutoDock Tools. This software identified the best ligand configurations and generated a PDBQT file. After optimization, the structures of the ligands Dapagliflozin and Etoricoxib, along with mutant PI3K, Akt, and mTOR signaling proteins, were loaded into AutoDock Tools. The docking process was then performed using the same software. The docking energy scores and binding interactions were analyzed with BIOVIA Discovery Studio, UCSF Chimera, and AutoDock Vina [50, 51].
2-5- Mapping Drug Interactions
2-5-1- (Combination Index- CI)
Compusyn, a computational simulator, calculated the combination index (CI) scores. This evaluation aimed to determine the potential for synergistic, additive, or antagonistic interactions among the mixture’s components. Concentration-effect curves illustrate the percentage of cells showing reduced growth concerning drug concentration, assessed after 24 and 72 hours of treatment.
CI values below 1 indicate synergy, values equal to 1 suggest additivity, and those above 1 indicate antagonism. The combination index was determined using Compusyn software (Biosoft, Ferguson, MO, USA) [52, 53].
2-5-2- (dose reduction index- DRI)
Compusyn, a computational simulator, calculated dose reduction index (DRI) scores. The DRI score indicates to what extent the concentration of each drug in a mixture can be decreased while still preserving its cytotoxic effectiveness.
A DRI score exceeding 1 signifies a beneficial decrease in concentration, whereas a score below 1 indicates an unfavorable reduction. The DRI analysis was performed using Compusyn software (Biosoft, Ferguson, MO, USA) calculations [52, 53].
2-6- Ethical approval
This study solely utilized in vitro cell line models, with no human participants or laboratory animals involved. All research methods followed institutional ethical standards for laboratory studies.
2-7- Statistical Analysis
The cytotoxicity assay results are shown as mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) was performed to evaluate the variation among the research groups. To analyze differences between specific groups, paired t-tests and LSD tests were utilized. Statistical analyses were conducted using SPSS version 20, with a significance level set at p < 0.05 [54].
The study employed lowercase and uppercase letters in data tables to differentiate between statistical groupings and significance levels. Means (averages) that share the same letter indicate no significant difference, while means with different letters are statistically significant. Uppercase letters are used for comparisons with the means of the rows, whereas lowercase letters compare the means of the columns. This method provides a clear and straightforward way to convey complex statistical findings without detailed explanations. Readers can quickly grasp which groups are similar or different based on the assigned letters.
3- Results
3-1- Cytotoxic assay
3-1-1(Dapagliflozin -etoricoxib) mixture cytotoxicity
The study findings demonstrate that the Dapagliflozin etoricoxib mixture inhibits cervical cancer growth, with the inhibition pattern mainly depending on the mixture’s concentration and incubation time. At the same time, the mixture shows a significantly lower cytotoxic effect on the NHF cell line compared to the cancer cell line, indicating a favorable safety profile and targeted toxicity against cancer cells (Table 1, 2).
| Cellular proliferation inhibition (mean ± SD) | ||||||
| Concentration (µg/ml) | HeLa cell line | NHF cell line | ||||
| 24 hr. | 72 hr. | P- value | 24 hr. | 72 hr. | P- value | |
| 0.1 | D 9.00 ± 0.000 | C 20.00 ± 1.000 | 0.158 | B 0.00 ± 0.000 | C 1.00 ± 1.000 | 0.158 |
| 1 | CD 16.00 ± 1.000 | C 25.00 ± 2.000 | 0.196 | B 1.00 ± 1.000 | BC 3.00 ± 2.000 | 0.196 |
| 10 | C 21.00 ± 1.000 | C 29.00 ± 2.000 | 0.018* | B 6.00 ± 1.000 | AB 11.00 ± 2.000 | 0.018* |
| 100 | B 47.00 ± 4.000 | B 52.00 ± 1.000 | 0.042* | AB 9.00 ± 4.000 | A 16.00 ± 1.000 | 0.042* |
| 1000 | A 71.00 ± 3.000 | A 85.00 ± 5.000 | 0.423 | A 16.00 ± 3.000 | A 19.00 ± 5.000 | 0.423 |
| LSD value | 8.46 | 9.62 | - | 7.11 | 8.53 | - |
| IC 50 | 125 µg/ml | 91.3 µg/ml | - | >1000 µg/ml | >1000 µg/ml | - |
*, significant at (P<0.05)
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | |||||
| 24 hr. | 72 hr. | |||||
| HeLa cell line | NHF cell line | P- value | HeLa cell line | NHF cell line | P- value | |
| 0.1 | D 9.00 ± 0.000 | B 0.00 ± 0.000 | 0.018* | C 20.00 ± 1.000 | C 1.00 ± 1.000 | 0.003* |
| 1 | CD 16.00 ± 1.000 | B 1.00 ± 1.000 | 0.003* | C 25.00 ± 2.000 | BC 3.00 ± 2.000 | 0.0001* |
| 10 | C 21.00 ± 1.000 | B 6.00 ± 1.000 | 0.0001* | C 29.00 ± 2.000 | AB 11.00 ± 2.000 | 0.0001* |
| 100 | B 47.00 ± 4.000 | AB 9.00 ± 4.000 | 0.0001* | B 52.00 ± 1.000 | A 16.00 ± 1.000 | 0.0001* |
| 1000 | A 71.00 ± 3.000 | A 16.00 ± 3.000 | 0.0001* | A 85.00 ± 5.000 | A 19.00 ± 5.000 | 0.0001* |
| LSD value | 8.46 | 7.11 | - | 9.62 | 8.53 | - |
| IC 50 | 125 µg/ml | >1000 µg/ml | - | 91.3 µg/ml | >1000 µg/ml | - |
*, significant at (P<0.05)
3-1-2- 5-FU cytotoxicity
5-FU was chosen as a positive control for comparison. Its cytotoxic effects on cancer cells (cervical cancer) and normal healthy cells (human adipose tissue) were assessed to determine its selective toxicity. Its effect on cancer cells showed a concentration-dependent pattern, while its cytotoxicity decreased over time, suggesting the development of resistance to 5-FU by the cancer cells. This is supported by the decline in the IC50 levels at 24- and 72-hour incubation.
At the same time, the cytotoxic effects of 5-FU on healthy normal cells varied depending on concentration and treatment duration. The level of cytotoxicity was comparable to its impact on the HeLa cancer cell line Table 3.
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | |||||
| HeLa cell line | NHF cell line | |||||
| 24 hr. | 72 hr. | P- value | 24 hr. | 72 hr. | P- value | |
| 0.1 | E 7.00 ± 2.000 | D 2.00 ± 1.000 | 0.018* | E 2.00 ± 2.000 | C 10.00 ± 5.000 | 0.062* |
| 1 | D 17.00 ± 2.000 | C 11.00 ± 1.000 | 0.010* | D 11.00 ± 1.000 | C 24.00 ± 4.000 | 0.005* |
| 10 | C 39.00 ± 1.000 | C 21.00 ± 4.000 | 0.002* | C 21.00 ± 2.000 | BC 36.00 ± 3.000 | 0.002* |
| 100 | B 71.00 ± 1.000 | B 47.00 ± 2.000 | 0.0001* | B 40.00 ± 3.000 | B 45.00 ± 5.000 | 0.212 |
| 1000 | A 89.00 ± 1.000 | A 59.00 ± 2.000 | 0.0001* | A 57.00 ± 2.000 | A 69.00 ± 2.000 | 0.002* |
| LSD value | 5.4 | 8.3 | - | 7.64 | 14.46 | - |
| IC 50 | 34.3 µg/ml | 250.1 µg/ml | - | 588.2 µg/ml | 208.8 µg/ml | - |
*, significant at (P<0.05)
3-1-3- Cytotoxic effects of mixture ingredients
We assessed the cytotoxicity of Dapagliflozin and etoricoxib to elucidate the mechanisms behind their combined toxicity. Furthermore, this investigation aids in analyzing the interactions between these medications, determining whether they exhibit synergistic, antagonistic, or additive effects.
3-1-3-1- Dapagliflozin Cytotoxicity
Dapagliflozin has demonstrated the capability to suppress cervical cancer growth; higher concentrations of Dapagliflozin and longer incubation times significantly increased cytotoxicity. The impact of concentration on cytotoxicity was more pronounced than that of incubation duration Table 4.
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | P- value | |
| 24 hr. | 72 hr. | ||
| 0.1 | D 0.00 ± 0.000 | D 1.00 ± 1.000 | 0.158 |
| 1 | C 11.00 ± 1.000 | C 13.00 ± 3.000 | 0.335 |
| 10 | C 14.00 ± 4.000 | C 17.00 ± 2.000 | 0.31 |
| 100 | B 34.00 ± 3.000 | B 39.00 ± 6.000 | 0.266 |
| 1000 | A 48.00 ± 4.000 | A 56.00 ± 1.000 | 0.028* |
| LSD value | 10.54 | 11.62 | |
| IC 50 | >1000 µg/ml | 812.9 µg/ml |
*, significant at (P<0.05)
3-1-3-2- Etoricoxib cytotoxicity
Etoricoxib has also been shown to inhibit the growth of cervical cancer cells. This effect appears to be directly proportional to the concentration of Etoricoxib, with no significant influence from time incubation Table 5.
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | P- value | |
| 24 hr. | 72 hr. | ||
| 0.1 | C 0.00 ± 0.000 | D 0.00 ± 0.000 | N. S |
| 1 | C 3.00 ± 3.000 | D 4.00 ± 1.000 | 0.613 |
| 10 | B 20.00 ± 5.000 | C 28.00 ± 2.000 | 0.062 |
| 100 | A 39.00 ± 2.000 | B 41.00 ± 1.000 | 0.196 |
| 1000 | A 41.00 ± 4.000 | A 49.00 ± 5.000 | 0.096 |
| LSD value | 11.96 | 9.06 | |
| IC 50 | >1000 µg/ml | >1000 µg/ml |
*, significant at (P<0.05)
3-1-4- Cytotoxicity comparison among Dapagliflozin, Etoricoxib, 5 FU, and the mixture
This comparison was conducted to investigate whether the mixture exhibited a greater cytotoxic effect than its ingredients. Further comparisons were made against 5-FU, the standard chemotherapy drug.
The findings from these comparisons indicated that, at most concentrations, the mixture exhibits a significantly higher cytotoxic effect than the individual ingredients, suggesting a synergistic cytotoxic interaction among the medications. Additionally, the cytotoxicity of the mixture was similar to that of 5-FU, except at the higher concentration after 72 hours of incubation, where the mixture exhibited a more pronounced cytotoxic effect than 5-FU Table 6, 7.
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | b LSD value | |||
| Dapagliflozin | Etoricoxib | combination | 5FH | ||
| 0.1 | D 0.00 ± 0.000 c | C 0.00 ± 0.000 c | D 9.00 ± 0.000 a | E 7.00 ± 2.000 bc | 8.42 |
| 1 | C 11.00 ± 1.000 ab | C 3.00 ± 3.000 b | CD 16.00 ± 1.000 a | D 17.00 ± 2.000 a | 10.32 |
| 10 | C 14.00 ± 4.000 b | B 20.00 ± 5.000 b | C 21.00 ± 1.000 b | C 39.00 ± 1.000 a | 12.34 |
| 100 | B 34.00 ± 3.000 cb | A 39.00 ± 2.000 b | B 47.00 ± 4.000 b | B 71.00 ± 1.000 a | 9.02 |
| 1000 | A 48.00 ± 4.000 b | A 41.00 ± 4.000 b | A 71.00 ± 3.000 a | A 89.00 ± 1.000 a | 10.98 |
| LSD value | 10.54 | 11.96 | 8.46 | 5.4 | |
| IC 50 | >1000 µg/ml | >1000 µg/ml | 125 µg/ml | 34.3 µg/ml |
*, significant at (P<0.05)
| Concentration (µg/ml) | Cellular proliferation inhibition (mean ± SD) | b LSD value | |||
| Dapagliflozin | Etoricoxib | combination | 5FH | ||
| 0.1 | D 1.00 ± 1.000 b | D 0.00 ± 0.000 b | C 20.00 ± 1.000 a | D 2.00 ± 1.000 b | 9.78 |
| 1 | C 13.00 ± 3.000 b | D 4.00 ± 1.000 c | C 25.00 ± 2.000 a | C 11.00 ± 1.000 bc | 8.42 |
| 10 | C 17.00 ± 2.000 b | C 28.00 ± 2.000 a | C 29.00 ± 2.000 a | C 21.00 ± 4.000 ab | 9.96 |
| 100 | B 39.00 ± 6.000 b | B 41.00 ± 1.000 ab | B 52.00 ± 1.000 a | B 47.00 ± 2.000 ab | 12.64 |
| 1000 | A 56.00 ± 1.000 b | A 49.00 ± 5.000 b | A 85.00 ± 5.000 a | A 59.00 ± 2.000 b | 11.76 |
| LSD value | 11.62 | 9.06 | 9.62 | 8.3 | |
| IC 50 | 812.9 µg/ml | >1000 µg/ml | 91.3 µg/ml | 250.1 µg/ml |
*, significant at (P<0.05)
3-2- Selective toxicity assay
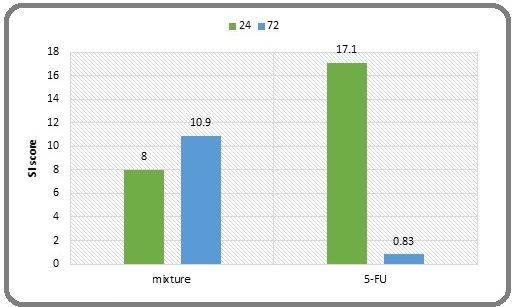
The selectivity index (SI) for the Dapagliflozin– Etoricoxib mixture was 8 and 10.9 after 24 and 72 hours, respectively. This indicates a greater selectivity of the mixture in targeting cancer cells compared to its effects on healthy cells. Notably, the results suggest that the SI increases with longer incubation periods. In contrast, the SI for 5-FU was 17.1 at 24 hours but significantly decreased to 0.83 at 72 hours. This decline reflects a reduced selectivity for cancer cells at the latter time point compared to the higher selectivity observed at 24 hours. This observation indicates a potential development of resistance in cancer cells to 5-FU as the incubation duration extends (Figure 1).
Figure 1. Comparison of the SI between the Mixture and 5-FU SI at 24 and 72 hours. (An SI above 1.0 indicates that a drug is more effective against tumor cells than against normal cells.).
3-3- Mixture medication Interaction Mapping
3-3-1-(Combination Index)
The CI score indicated a varied pattern of interactions among the mixture ingredients. After 24 hours of incubation, concentrations of 0.1 and 1 μg/ml displayed very strong synergism, 10 μg/ml exhibited strong synergism, 100 μg/ml showed synergism, whereas 1000 μg/ml reflected moderate synergism.
After 72 hours of incubation, the results indicated that concentrations of 0.1 and 1 μg/ml exhibited very strong synergism, while concentrations of 10 and 1000 μg/ml displayed strong synergism. In contrast, 100 μg/ml reflected synergism (Table 8, 9) (Figure 2).
| Concentration μg/ml | Combination pattern | DRI score | |||||
| Dapagliflozin | Etoricoxib | Mix | ratio | CI score | Dapagliflozin | Etoricoxib | |
| 0.05 | 0.05 | 0.1 | 1:01 | 0.00876 | Very Strong Synergism | 199.265* | 267.094* |
| 0.5 | 0.5 | 1 | 0.03313 | Very Strong Synergism | 54.2034* | 68.1374* | |
| 5 | 5 | 10 | 0.20206 | Strong Synergism | 9.01647* | 10.9704* | |
| 50 | 50 | 100 | 0.33921 | Synergism | 5.67436* | 6.13572* | |
| 500 | 500 | 1000 | 0.75573 | Moderate Synergism | 2.67489* | 2.61858* |
The CI (Combination Index) and DRI (Dose Reduction Index) values were computed utilizing Compusyn software. A CI exceeding 1 signifies antagonism, a CI of 1 denotes an additive effect, and a CI below 1 suggests synergism. A DRI greater than one is associated with decreased toxicity. (*) indicates a beneficial reduction in the effective cytotoxic concentration. [55]
| Concentration μg/ml | CI score | Combination pattern | DRI score | ||||
| Dapagliflozin | Etoricoxib | Mix | ratio | Dapagliflozin | Etoricoxib | ||
| 0.05 | 0.05 | 0.1 | 1:01 | 0.00428 | Very Strong Synergism | 350.629* | 700.051* |
| 0.5 | 0.5 | 1 | 0.0253 | Very Strong Synergism | 63.5931* | 104.467* | |
| 5 | 5 | 10 | 0.1754 | Strong Synergism | 9.68428* | 13.8610* | |
| 50 | 50 | 100 | 0.32283 | Synergism | 7.29055* | 5.38596* | |
| 500 | 500 | 1000 | 0.2304 | Strong Synergism | 22.3791* | 5.38446* |
The CI (Combination Index) and DRI (Dose Reduction Index) values were computed utilizing Compusyn software. A CI exceeding 1 signifies antagonism, a CI of 1 denotes an additive effect, and a CI below 1 suggests synergism. A DRI greater than one is associated with decreased toxicity. (*) indicates a beneficial reduction in the effective cytotoxic concentration. [55]
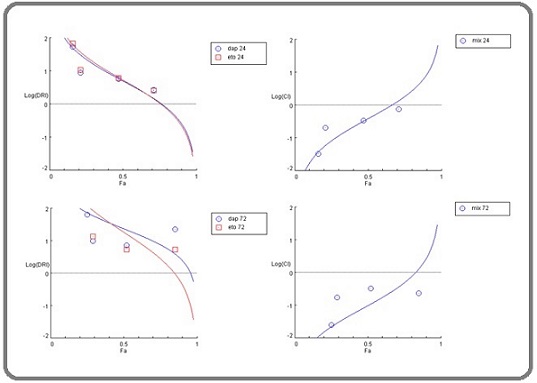
Figure 2. Log the combination index (CI) on the right and the dose reduction index (DRI) on the left for the mixture after a 24-hour incubation period (above) and a 72-hour incubation period (below). dap: Dapagliflozin, eto: etoricoxib, CI: combination index, DRI: dose reduction index.
3-3-2-(dose reduction index)
Throughout each incubation period and across all concentrations, the DRI score consistently exceeded 1. This indicates that the concentration of each ingredient in the mixture causing significant cytotoxicity was lower than the significant cytotoxic concentrations of each ingredient used individually. This reduction in concentrations suggests a decreased likelihood of side effects from the mixture, especially when compared to the risks associated with each drug when used alone (Table 8, 9) (Figure 2).
3-4- Molecular docking studies
A molecular docking simulation was used to test the interaction between the mixture components (Dapagliflozin and etoricoxib) and mutant signal proteins of the PI3K-Akt-mTOR pathway, utilizing protein data bank codes 6oac, 6s9k, and 4jsv for each protein, respectively. A mutant structure was selected for docking studies because it represents a constitutively active, disease-relevant conformational state, which is a primary target for therapeutic inhibition in oncology. The study employed AutoDock Tools version 1.5.7, BIOVIA Discovery Studio, UCSF Chimera, and AutoDock Vina [56].
3-4-1- Docking with mutant PI3K signal protein
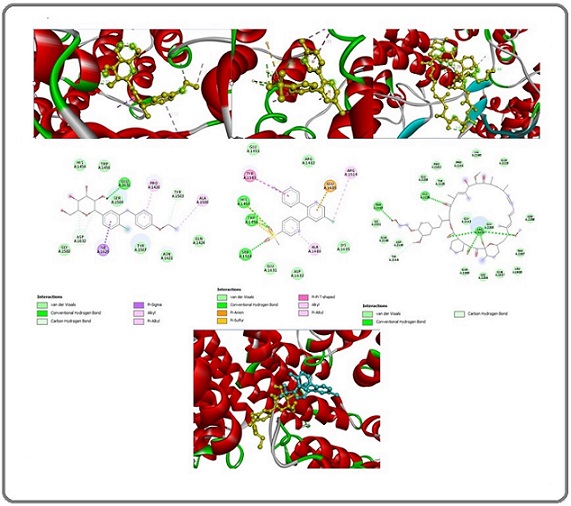
The chemical docking study findings revealed that Dapagliflozin interacted with the mutant PI3K signal protein, achieving a docking score of -8.0 kcal/mol. Molecular docking analysis shows three “conventional hydrogen bonds” with the amino acid residues TYR A:836, CYS A:838, and GLN A:630 at distances of 2.82 Å, 2.59 Å, and 2.62 Å, respectively. Two “alkyl bonds” occur with residues ARG A:662 and PRO A:757 at 4.38 Å and 4.36 Å, respectively. One “pi-sigma bond” forms with GLN A:630 at 3.90 Å, and an additional “alkyl bond” forms with ARG A:818 at 4.80 Å (Figure 3).
Figure 3. Binding Site Structures in 2D (middle image) and 3D (top image) for Mutant PI3K with (Dapagliflozin, etoricoxib, and alpelisib) from Left to Right. The lower 3D structure shows the binding sites of Dapagliflozin (yellow) and etoricoxib (blue) in conjunction with the mutant PI3K.
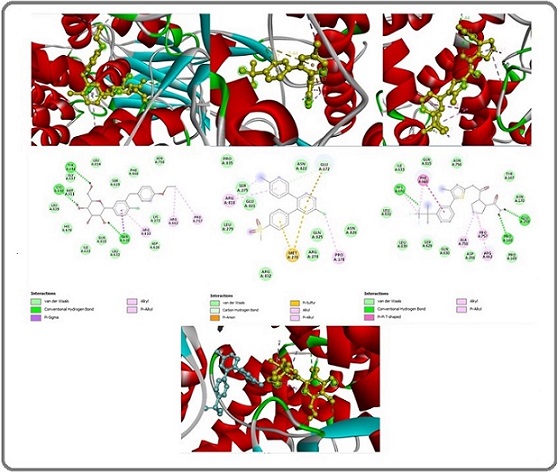
Another ingredient in the mixture, etoricoxib, demonstrated the ability to interact with the mutant PI3K signaling protein, achieving a docking score of -8.0 kcal/ mol. Molecular docking analysis shows one “carbon hydrogen bond” with the amino acid residue GLU A:172 at a distance of 3.60 Å, one “pi-anion bond” with the amino acid residue GLU A:172 at a distance of 3.87 Å, two “pi- sulfur bonds” with two MET A:278 residues at distances of 5.43 Å and 3.78 Å, respectively. Additionally, two “alkyl bonds” form with ARG A:818 and PRO A:178 residues at distances of 3.83 Å and 4.31 Å, respectively. Lastly, a “pi-alkyl bond” forms with the ARG A:818 residue at a distance of 5.23 Å (Figure 3).
Docking sites for each mixture of ingredients with PI3K showed diversity, indicating that the mixture ingredients act synergistically in terms of targeting PI3K. (Figure 3).
For comparative purposes, a molecular docking study assessed the interaction between alpelisib, a “ PI3K inhibitor.” [57]. A mutant PI3K signal protein, resulting in a docking score of -9.1 kcal/mol for binding and presenting. Three “conventional hydrogen bond” constraints with HIS A:670, GLU A:295, and PRO A:168 amino acid residues at distances of 2.48 Å, 2.10 Å, and 2.44 Å, respectively. Two “pi-alkyl bonds” constrict with PHE A:666 and HIS A:670 amino acid residues at distances of 4.36 Å and 4.82 Å, respectively. And finally, with One “pi-pi-T-shaped bond” constricts with PHE A:666 amino acid residues at a distance of 5.14 Å (Figure 3).
3-4-2- Docking with mutant AKt signal protein
The molecular docking study results of the interaction between the mutant AKt signal protein and Dapagliflozin showed a docking score of -6.7 kcal/mol. Molecular docking analysis revealed one “conventional hydrogen bond” with the amino acid residue GLU A:163 at a distance of 2.72 Å. Three “carbon-hydrogen bonds” occur with two residues of ASP A:1632 and one residue of TYR A:1583 at 3.51 Å, 3.19 Å, and 3.60 Å, respectively. One “pi-sigma bond” with the amino acid residue ILE A:1629 at a distance of 3.63 Å. One “alkyl bond” with the amino acid residue ALA A:1586 at a distance of 3.80 Å. And finally, one “pi-alkyl bond” with the amino acid residue PRO A:1426 at a distance of 4.74 Å (Figure 4).
Figure 4. Binding Site Structures in 2D (middle image) and 3D (top image) for Mutant AKt with (Dapagliflozin, etoricoxib, and capivasertib) from Left to Right. The lower 3D structure shows the binding sites of Dapagliflozin (yellow) and etoricoxib (blue) in conjunction with the mutant AKt.
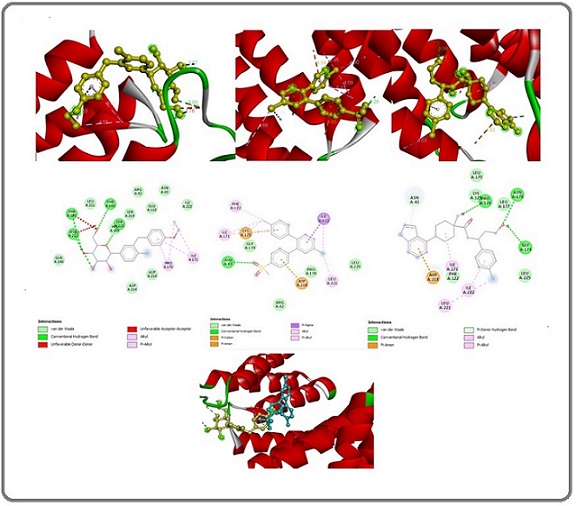
Regarding the interaction of etoricoxib with the mutant AKt signaling protein, the docking score was -6.8 kcal/mol. Molecular docking analysis revealed three “conventional hydrogen bonds” with the amino acid residues HIS A:1454, TRP A:1456, and SER A:1584 at distances of 2.73 Å, 2.13 Å, and 2.22 Å, respectively. One “pi-anion bond” occurs with the amino acid residue GLU A:1485 at 3.65 Å. Three “pi-sulfur bonds” occur with one amino acid residue HIS A:1454 and two amino acid residues of TRP A:1456 at distances of 4.84 Å, 5.29 Å, and 5.47 Å, respectively. One “pi-pi-shaped bond” occurs with the amino acid residue TYR A:1583 at 5.30 Å. One “alkyl bond” occurs with the amino acid residue ARG A:1514 at 5.00 Å. Lastly, two “pi-alkyl bonds” occur with residues TYR A:1583 and ALA A:1486 at distances of 4.72 Å and 5.15 Å, respectively (Figure 4).
Docking sites for each ingredient mixture with AKt exhibited variability, suggesting that the mixture ingredients interact synergistically in their targeting mechanisms (Figure 4).
For comparison, a molecular docking study evaluated the interaction between capivasertib (an AKT inhibitor) [58] And a mutant PI3K signaling protein, resulting in a docking score of -6.7 kcal/mol for binding. The study showed three “conventional hydrogen bond” constraints with ASN A:178, GLY A:174, and PRO A:170 amino acid residues at distances of 3.06 Å, 2.19 Å, and 2.62 Å, respectively. It also identified one “pi-alkyl bond” constraint with ASP A:218 at 4.61 Å, one “pi-donor hydrogen bond” with ASN A:43 at 3.16 Å, two “alkyl bond” constraints with ILE A:171 and LEU A:221 at 4.68 Å and 4.62 Å, respectively, and finally, one “pi-alkyl bond” with ILE A:222 at 4.98 Å (Figure 4).
3-4-3- Docking with mutant mTOR signal protein
The molecular docking study results of the interaction between Dapagliflozin and the mutant mTOR signal protein yield a docking score of -7.3 kcal/mol. Molecular docking analysis revealed one “conventional hydrogen bond” with the amino acid residues GLU A:163, at distances of 2.72 Å. Three “carbon-hydrogen bonds” occur with two residues of ASP A:1632 and one residue of TYR A:1583 at 3.51 Å, 3.19 Å, and 3.60 Å, respectively. one “pi-sigma bond” with the amino acid residue ILE A:1629 at a distance of 3.63 Å. one “Alkyl bond” with the amino acid residue ALA A:1586 at a distance of 3.80 Å. And finally, one “pi-alkyl bonds” occurs with residues PRO A:1426 at 4.74 Å (Figure 5).
Figure 5. Binding Site Structures in 2D (middle image) and 3D (top image) for Mutant mTOR with (Dapagliflozin, etoricoxib, and Everolimus) from Left to Right. The lower 3D structure shows the binding sites of Dapagliflozin (yellow) and etoricoxib (blue) in conjunction with the mutant mTOR.
Regarding the interaction of etoricoxib with the mutant mTOR signaling protein, the results showed a docking score of -6.8 kcal/mol. Molecular docking analysis revealed three “conventional hydrogen bonds” with the amino acid residues HIS A:1454, TYR A:1456, and SER A:1584 at distances of 2.73 Å, 2.13 Å, and 2.22 Å, respectively. One “pi-anion bond” occurs with the amino acid residue GLU A:1485 at 3.65 Å. Three “pi-sulfur bonds” occur with the amino acid residues HIS A:1454 and TRP A:1456 at 4.84 Å, 5.29 Å, and 5.47 Å, respectively. One “pi-pi-shaped bond” occurs with the amino acid residue TYR A:1583 at 5.30 Å. One “alkyl bond” occurs with the amino acid residue ARG A:1514 at 5.00 Å. Lastly, two “pi-alkyl bonds” occur with residues TYR A:1583 and ALA A:1486 at 4.72 Å and 5.15 Å, respectively. Figure 5 Based on the interaction patterns of each mixture ingredient with mTOR, we proposed that the combination exerts its effects through a complementary mechanism, indicating a synergistic interaction among the components (Figure 5).
For comparison purposes, a molecular docking study evaluated the interaction between Everolimus (an mTOR inhibitor) [59]. The mutant mTOR signaling protein was used, resulting in a docking score of -9.7 kcal/mol for the binding interaction. The study showed seven “conventional hydrogen bond” constraints with five ARG A:2224, one ALA A:2226, and one THR A:2143 amino acid residues at distances of 2.68 Å, 2.31 Å, 2.39 Å, 1.94 Å, 2.56 Å, 2.51 Å, and 2.44 Å, respectively. And finally, one “carbon hydrogen bond” constraint was identified with TYR A:2144 at 3.45 Å (Figure 5).
To clarify the mixture’s effectiveness in targeting PI3K-Akt-mTOR signaling proteins, a comparison of its docking score with standard medications targeting the same pathway was performed (Table 10).
| Molecular target | Dapagliflozin | Etoricoxib | Alpelisib | Capivasertib | Everolimus |
| PI3K | -8 | -8 | -9.1 | - | - |
| AKt | -6.7 | -6.6 | - | -6.7 | - |
| mTOR | -7.3 | -6.8 | - | - | -9.7 |
PI3K, phosphoinositide 3-kinase; Akt, protein kinase B; mTOR, mammalian target of rapamycin. Docking scores (in kcal/mol) indicate the predicted binding affinity, with more negative values representing stronger binding. The scores for the standard inhibitors (alpelisib for PI3K, capivasertib for Akt, and Everolimus for mTOR) are included for comparison and were generated using the same molecular docking method and mutant protein structures (PDB: 6OAC, 6S9K, 4JSV) as the test compounds. A dash (—) shows that the docking simulation was not performed for that specific drug-target pair, as it was outside the scope of this comparative analysis.
3-5- Histopathological features of the study cell lines
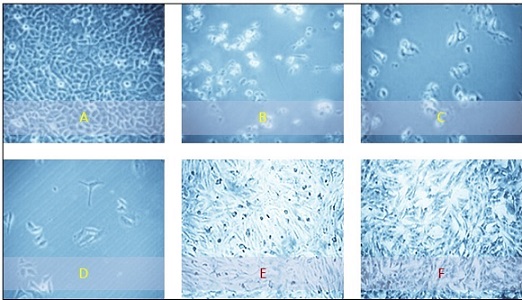
Figure 6 shows the morphological changes in the HeLa and NHF cell lines after 72 hours of treatment.
Figure 6. The Morphological Characteristics of the Study Cell Lines Seen through an Inverted Microscope (400X). (A) HeLa cancer cells without any treatment.(B) Hela cells treated with 100 µg/ml of Dapagliflozin for 72 hours. (C) Hela cells exposed to 1000 µg/ml of etoricoxib for 72 hours. (D) Hela cells treated with 1000 µg/ml of a mixture for 72 hours. (E) Untreated human-derived adipose tissue (NHF cell line). (F) NHF normal cells were exposed to a 1000 µg/ml concentration of a rivaroxaban–etoricoxib mixture for 72 hours .
4- Discussion
This research explores the potential of repurposing existing non-cancer medications as effective and safer alternatives in cancer therapy. Specifically, it examines the anticancer properties of a mixture of Dapagliflozin (a sodium-glucose cotransporter-2 (SGLT2) inhibitor) and etoricoxib (a cyclooxygenase-2 (COX-2) inhibitor). The rationale for selecting these agents is supported by numerous prior studies demonstrating that each medication possesses anticancer effects. Furthermore, a comprehensive evaluation of each drug’s pharmacokinetics and safety profiles has been conducted to ensure their viability in a combined treatment approach.
The results of the MTT cytotoxicity assay indicated that the combination of Dapagliflozin and etoricoxib effectively suppressed the growth of cervical cancer cells while demonstrating decreased cytotoxicity toward normal cell lines. This combination exhibited anticancer efficacy comparable to that of 5-fluorouracil (5FU) and was more effective than the individual effects of each drug when used separately.
Furthermore, the CI score suggests that the two medications are effective when used synergistically. Additionally, the DRI score shows that using this mixture decreases the likelihood of side effects compared to using each drug separately. The safety of the mixture is further supported by its favorable SI score, which demonstrates that it specifically targets cancer cells while sparing healthy cells. In contrast, 5FU showed a lower SI score at 72 hours compared to 24 hours, indicating that cancer cells developed less resistance to the mixture over time than to 5FU alone.
The anticancer mechanism of the mixture can be examined from two perspectives: first, by examining the proposed anticancer mechanisms of each ingredient based on previous research, and second, by exploring the anticancer mechanism revealed through our molecular docking study.
Recent findings support that SGLT2 inhibitors could offer benefits beyond managing diabetes. Research conducted both in vivo and in vitro has shown that dapagliflozin can reduce cancer cell growth and promote programmed cell death, especially in models of breast and prostate cancer [24]. The observed effects can be attributed to the drug’s capacity to disrupt glucose metabolism in cancer cells, thus impairing their energy supply and inhibiting growth [25]. Furthermore, SGLT2 inhibitors appear to influence key signaling pathways, including the PI3K/AKT/mTOR pathway, which is crucial for tumor survival and progression [26]. These mechanisms suggest that dapagliflozin could serve as an adjunctive therapy in conjunction with conventional cancer treatments, potentially enhancing their efficacy while mitigating metabolic side effects. However, the variable outcomes observed in particular malignancies, such as hepatocellular carcinoma, indicate that the drug’s impact may be context- dependent [26].
Additionally, our findings regarding the anticancer effects of etoricoxib align with previous research demonstrating its antiproliferative and pro-apoptotic properties in various cancer cell lines, including those from colorectal, breast, and lung cancers [30]. The observed reduction in tumor growth may be attributed to etoricoxib’s ability to inhibit the synthesis of prostaglandin E2 (PGE2), a crucial element in the pathogenesis of inflammation-related cancers. This inhibition could play a significant role in mitigating the inflammatory processes that contribute to tumor progression [31]. Etoricoxib inhibits COX-2-related PGE2 production and disrupts critical oncogenic pathways, including NF-κB and Wnt/β-catenin signaling. These pathways are well-established for promoting cell survival, angiogenesis, and metastasis, highlighting the potential of etoricoxib in targeting tumor progression [32]. Additionally, our findings suggest that etoricoxib enhances the effectiveness of standard chemotherapy drugs, likely by reducing drug resistance associated with inflammatory tumor microenvironments [33]. However, the efficacy of various anticancer agents appears to vary based on the tumor’s molecular characteristics, particularly in relation to COX-2 expression levels. Moreover, cancers with high COX-2 expression, such as colorectal cancer, tend to respond more favorably to specific treatments than those with low COX-2 activity. This differential response highlights the importance of personalized medicine approaches, which consider tumor biology when developing treatment strategies [34, 60, 61].
In addition to the previously mentioned anticancer mechanisms of each ingredient, this study investigates a new anticancer mechanism for both Dapagliflozin and Etoricoxib. The results from molecular docking suggest their capacity to block three key signaling proteins within the PI3K/Akt/mTOR pathway, exhibiting varying levels of interaction. A molecular docking study was conducted to evaluate the ability of Dapagliflozin to interact with the PI3K/Akt/mTOR signal protein, yielding docking scores of -8.0, -6.7, and -7.3 kcal/mol, respectively. At the same time, the etoricoxib docking scores were -8.0, -6.6, and
-6.8 kcal/mol, respectively.
In our study, we focused on investigating the capability of the mixture ingredient to target the PI3K/ Akt/mTOR pathway, given its critical role in cancer progression through the regulation of cell survival, proliferation, and metabolism [62]. Hyperactivation of this pathway, frequently observed in various malignancies, has been shown to enhance tumor proliferation and contribute to resistance against therapeutic interventions. This phenomenon underscores the critical role of this signaling cascade in cancer progression and highlights the potential for targeting this pathway in the development of more effective treatment strategies [63]. Pharmacological inhibition of the PI3K/Akt/mTOR signaling pathway has emerged as a promising therapeutic strategy in oncology, particularly for cancers driven by mutations in this pathway. Inhibitors such as alpelisib, which is specific to PI3Kα, and Everolimus, an mTOR inhibitor, have shown clinical efficacy in various settings. These agents provide a targeted approach, potentially leading to improved outcomes in patients with specific genetic alterations that activate the PI3K/Akt/mTOR signaling cascade. As research progresses, understanding the nuanced effects and optimizing the use of these inhibitors remains critical for advancing treatment strategies in cancer care [64]. Pathway redundancy and feedback mechanisms often limit the efficacy of single-agent therapies, underscoring the need for combination therapies that simultaneously target multiple nodes. Such approaches are crucial for achieving sustained therapeutic responses [65]. The PI3K/Akt/mTOR pathway is frequently dysregulated in cervical cancer, driving tumor progression and treatment resistance [66, 67].
Our molecular docking results indicate that dapagliflozin and etoricoxib have significant binding affinities (-8.0 kcal/mol) to mutant PI3K, although they interact through different mechanisms. Dapagliflozin establishes multiple hydrogen bonds with catalytic residues (TYR836, CYS838) and engages in hydrophobic interactions. In contrast, etoricoxib forms unique pi-anion and pi-sulfur bonds with residues GLU172 and MET278. This complementary binding profile suggests a potential for synergistic inhibition of PI3K. While alpelisib (a PI3K inhibitor) demonstrates a higher binding affinity (-9.1 kcal/ mol) through conventional hydrogen bonds with essential catalytic residues (HIS670, GLU295), the multi-target approach of combining dapagliflozin and etoricoxib may provide advantages in overcoming drug resistance.
Furthermore, our molecular docking studies show that both dapagliflozin (-6.7 kcal/mol) and etoricoxib (-6.8 kcal/mol) have similar binding affinities to the mutant AKt, although they utilize different interaction profiles. Dapagliflozin primarily interacts with GLU163 through hydrogen bonds and hydrophobic interactions, particularly alkyl/pi-alkyl bonds. Meanwhile, etoricoxib forms several conventional hydrogen bonds with HIS1454 and TRP1456, as well as unique pi-anion and pi-sulfur interactions. This difference in binding suggests the possibility of synergistic inhibition of AKt signaling. Interestingly, capivasertib (an Akt inhibitor) shows a comparable binding affinity of -6.7 kcal/mol but engages with different catalytic residues, specifically ASN178 and GLY174. The varied interaction patterns of the mixture ingredients support their potential as modulators of the AKt pathway.
The molecular docking results of the present study indicate that dapagliflozin has a stronger binding affinity to the mutant mTOR (-7.3 kcal/mol) compared to etoricoxib (-6.8 kcal/mol). This suggests that dapagliflozin may be a more effective modulator of the mTOR pathway. Dapagliflozin’s binding is stabilized through hydrogen bonding with GLU163 and several hydrophobic interactions. In contrast, etoricoxib forms an extensive network of hydrogen bonds and unique pi-anion and pi-sulfur interactions. This complementary binding profile indicates the potential for a synergistic effect between the two compounds. However, the significantly higher affinity of Everolimus (-9.7 kcal/mol) is notable, as it achieves this through seven conventional hydrogen bonds with key residues, underscoring its superior binding efficiency compared to other mTOR inhibitors. These findings suggest that while both dapagliflozin and etoricoxib demonstrate promising mTOR binding capabilities.
The PI3K/Akt/mTOR signal proteins are integral to various cellular functions, including growth, proliferation, and survival, making them a pivotal focus in cancer research. Abnormal activation of this pathway is frequently associated with numerous cancers, positioning its components as promising therapeutic targets for these diseases. A variety of clinical trials have been conducted to identify and evaluate potential inhibitors that specifically target these signaling proteins, aiming to develop effective treatment strategies that could improve patient outcomes in oncological settings, as Alpelisib (PI3Kα inhibitor) [64] Capivasertib (AKT inhibitor) [68] Everolimus (mTOR inhibitor) [69], and Sapanisertib (dual mTORC1/2 inhibitor) [70].
Identifying various inhibitors targeting the PI3K/ Akt/mTOR signaling pathway has garnered significant attention in therapeutic settings. However, several challenges often hinder the clinical success of these agents when used alone. Major issues include drug resistance, the development of metabolic toxicities, and the activation of compensatory signaling pathways, all of which diminish the effectiveness of these inhibitors. Consequently, there is a continuing need to explore alternative treatment strategies and combination therapies to improve patient outcomes [71, 72]. Strategies that incorporate multiple approaches are being studied to address these issues [73]. While our molecular docking data indicate that Dapagliflozin and Etoricoxib may effectively bind to key components of the PI3K/Akt/mTOR pathway, this study has notable limitations, primarily the absence of experimental validation at the protein level, such as Western Blot analysis of phosphorylated Akt (p-Akt), mTOR (p-mTOR), and their downstream effectors (e.g., p-p70S6K, p-4E-BP1). This represents a significant gap. Consequently, the docking results should be interpreted as providing robust support for the observed synergistic cytotoxicity. The primary contribution of this work is the demonstration of a pronounced in vitro synergistic effect and a favorable safety profile for the drug combination, supported by a computationally predicted mechanistic rationale. Future investigations are necessary to validate this proposed mechanism experimentally. Our subsequent step, contingent upon funding, is to perform Western Blot analysis on HeLa cells treated with this drug combination to confirm pathway inhibition.
In conclusion, this study aimed to identify an effective and safe anticancer option by repurposing a combination of two marketed medications (Dapagliflozin and etoricoxib) for malignant cervical cancer treatment. MTT assay findings indicate that the mixture of Dapagliflozin and etoricoxib significantly inhibits cervical cancer growth compared to 5FU, Dapagliflozin, and etoricoxib cytotoxicity. Dapagliflozin and etoricoxib exhibit synergistic effects when used in combination, as indicated by the combination index score. Regarding safety, the effective cytotoxic concentrations of the drugs in the mixture were lower than when the drugs were used individually, indicating that the mixture is less likely to cause adverse effects. The mixture exhibits a favorable selectivity index, indicating that it selectively targets cancer cells over healthy cells, particularly at 72 hours of incubation.
The study explores a novel anticancer mechanism of the mixture, demonstrating dual targeting of the PI3K/ Akt/mTOR pathway: PI3K/Akt/mTOR signal proteins are targeted by Dapagliflozin with molecular docking scores of -8, -6.7, and -7.3 kcal/mol, respectively, while etoricoxib exhibits -8, -6.7, and -7.3 kcal/mol, respectively. This proposed mechanism explains the potential anticancer effects of the mixture and highlights the synergistic interactions that occur between its components.
Study outcomes highlight the potential for integrating diverse therapeutic approaches, emphasizing the importance of understanding the multifaceted interactions within the treatment framework to enhance efficacy. Expanding on these findings, further research could investigate the specific pathways involved and their implications for clinical applications in cancer therapy.
Author Contributions
Design and development: Solafa Rabi Salih, Aqeela Hayder Majeed, Kawakib Majid Hussein.
Gathering and organizing data: Youssef Shakuri Yasin, Azal Hamoody Jumaa.
Data analysis/interpretation: Azal Hamoody, Aqeela Hayder Majeed, Kawakib Majid Hussein.
Article composition: Solafa Rabi Salih, Aqeela Hayder Majeed ,Azal Hamoody.
Critique the essay for significant ideas: Solafa Rabi Salih ,Azal Hamoody.
Statistical analysis expertise: Aqeela Hayder Majeed, Kawakib Majid Hussein, Youssef Shakuri Yasin,
Ultimate article endorsement and guarantee: Solafa Rabi Salih, Aqeela Hayder Majeed, Kawakib Majid Hussein.
Acknowledgements
The research team would like to thank the researchers and instructional staff at ICMGR/Al-Mustansiriyah University and the Iraqi National Cancer Research Centre/ University of Baghdad for their vital support during this study. I also sincerely appreciate the quality control team at the Samarra Pharmaceutical Factory for supplying the drugs used in this research.
Financial support and sponsorship
Self-funded
Conflicts of interest:
The authors state that they have no conflicts of interest.
Declaration of Generative AI and AI-assisted technologies in the writing process:
The authors state that this work does not employ any generative AI or AI-assisted tools.
Abbreviations
(ICCMGR), The Iraqi Centre for Cancer and Medical Genetics Research. MTT, 3-(4,5-dimethylthiazol-2-yl)- 2,5-diphenyltetrazolium bromide stain. MEM, Minimum Essential Medium. SAS, Statistical Analysis System. SD, Standard deviation. LSD, Least Significant Difference. DRI, dose reduction index. CI, combination index. WHA, wound healing assay. NHF cell line, human-derived adipose tissue cell line. EMT, epithelial-mesenchymal transition. MMPs, matrix metalloprotease enzyme. TNKs, Tankyrase enzyme. ECM, extracellular matrix.
References
- Incidence and risk factors of vte in patients with cervical cancer using the korean national health insurance data Yuk J.-S., Lee B., Kim M.H., Kim K., Seo Y.-S., Hwang S.O.. Scientific Reports.2021. CrossRef
- Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis Arbyn M., Weiderpass E., Bruni L., Sanjosé S., Saraiya M., Ferlay J.. The Lancet Global Health.2020. CrossRef
- The effect of twist expression on the development ofcervical carcinoma in a group of iraqi women infected with hpv Khashman B.M., Abdulla K.N., Karim S., Alhashimi S., Mohammed M.L., Sarah N.A.. Biochem Cell Arch.2019;19(2).
- Cervical cancer screening programmes and age-specific coverage estimates for 202 countries and territories worldwide: A review and synthetic analysis Bruni L., Serrano B., Roura E., Alemany L., Cowan M., Herrero R.. The Lancet Global Health.2022. CrossRef
- Cervical cancer Cohen P.A., Jhingran A., Oaknin A., Denny L.. The Lancet.2019;393(10167). CrossRef
- Integrated genomic and molecular characterization of cervical cancer Network C.G.A.R.. Nature.2017. CrossRef
- Physiological scrutiny to appraise a flavonol versus statins Hashim W.S., Yasin Y.S., Jumaa A.H., Al-Zuhairi M.I., Abdulkareem A.H.. Biomedical and Pharmacology Journal.2023. CrossRef
- Most common risk factors distribution for cervical cancer Abdulla K.N., Abid S.J., Salih Alheshimi, SJ Al-Attar. Revista Latinoamericana de Hipertension.2024. CrossRef
- The prevalence of cesarean section in misan province and it’s indicating factors Alheshimi S.J., Majeed A.H., Abdulla K.N., Rassme H.A., Fadhil N.K., Fawzi H.A.. EXECUTIVE EDITOR.2019. CrossRef
- Novel therapeutics for recurrent cervical cancer: Moving towards personalized therapy Cohen AC LIC . Drugs.2020. CrossRef
- Evaluation of quality of life for women with breast cancer Khalifa M.F., Ghadhban A.G.Z.A., Hade I.M., Ali M.M.. Scripta Medica.2024.
- Changing paradigms in the systemic treatment of advanced cervical cancer Pfaendler K.S., Tewari K.S.. American journal of obstetrics and gynecology.2016. CrossRef
- Esomeprazole and amygdalin combination cytotoxic effect on human cervical cancer cell line (hela cancer cell line Jumaa A.H., Al Uboody W.S.H., Hady A.M.. Journal of Pharmaceutical Sciences and Research.2018.
- The effect of esomeprazole on cell line human cervical cancer Jumaa A.H., Jarad A.S., Al Uboody W.S.H.. Medico-Legal Update.2020.
- The cytotoxic effect of ciprofloxacin laetrile combination on esophageal cancer cell line Jumaa A.H., Abdulkareem A.H., Yasin Y.S.. Asian Pacific Journal of Cancer Prevention: APJCP.2024. CrossRef
- Antiproliferative impact of linagliptin on the cervical cancer cell line Said A.M., Abdulla K.N., Ahmed N.H., Yasin Y.S.. Asian Pacific Journal of Cancer Prevention: APJCP.2024.
- Synergistic antiproliferative effect of linagliptin-metformin combination on the growth of hela cancer cell line Ahmed A., Nihad A., Sabreen G., Youssef Y., Azal J.. Journal of Cancer Research Updates.2025.
- The cytotoxic effect of ephedra transitoria on hela cancer cell line: Bio-engineering Arean AG , Jumaa AH , Hashim WS , Mohamed AA , Yasin YS . .
- The cytotoxic effect of ethanolic extract of cnicus benedictus l. Flowers on the murine mammary adenocarcinoma cancer cell line amn-3 YS YA , AH J, WS H, YI K. Biochemical & Cellular Archives.2020.
- Laetrile and methotrexate: A dual-drug approach to inhibiting cervical cancer cell proliferation Abdulla K.N., Khudhur R.K., Said A.M., Jumaa A.H., Yasin Y.S.. Journal of Cancer Research Updates.2025.
- Scrutiny of the co-cytotoxic impact of metformin-omeprazole on the cervical cancer cell line and their aptitude to target heat shock 60 Khudhur R.K., Yahiya Y.I., Majeed A.H., Yasin Y.S., Jumaa A.H.. Asian Pacific Journal of Cancer Prevention.2025. CrossRef
- Effect of laetrile vinblastine combination on the proliferation of the hela cancer cell line Yasin Y.S., Jumaa A.H., Jabbar S., Abdulkareem A.H.. Asian Pacific Journal of Cancer Prevention: APJCP.2023. CrossRef
- Impact of esomeprazole, ciprofloxacin and their combination on cervical cancer cell line proliferation: A focus on heat shock protein 70 modulation Salih A, KN K, A Y, YS J. Asian Pacific Journal of Cancer Prevention.2025. CrossRef
- Antidiabetic agents as potential cytotoxic candidates for cancer therapy Alnaser RI , Alassaf FA , Abed MN . . CrossRef
- Exploring the role of sglt2 inhibitors in cancer: Mechanisms of action and therapeutic opportunities Pandey A, Alcaraz Jr M, Saggese P, Soto A, Gomez E, Jaldu S, et al . Cancers.2025. CrossRef
- Canagliflozin, a sglt-2 inhibitor, relieves er stress, modulates autophagy and induces apoptosis in irradiated hepg2 cells: Signal transduction between pi3k/akt/gsk-3β/mtor and wnt/β-catenin pathways;: In vitro Abdel-Rafei M.K., Thabet N.M., Rashed L.A., Moustafa E.M.. Journal of Cancer Research and Therapeutics.2021. CrossRef
- Dapagliflozin–structure, synthesis, and new indications Balkanski S.. Pharmacia.2021.
- Acid degradation study of dapagliflozin: Structural characterization of dapagliflozin and its degradation products using advanced analytical techniques and in silico toxicity prediction Bhuma D.R., Maddala V.K.S., Raghupathi J.K., Salakolusu S., Ranga M., Kaliyapermal M.. Separation Science Plus.2025;8(1).
- Design of improved antidiabetic drugs: A journey from single to multitarget agents Tassopoulou V.P., Tzara A., Kourounakis A.P.. ChemMedChem.2022. CrossRef
- Chemopreventive role of etoricoxib (mk-0663) in experimental colon cancer: Induction of mitochondrial proapoptotic factors Tanwar L., Vaish V., Sanyal S.N.. European Journal of Cancer Prevention.2010. CrossRef
- Selective cyclooxygenase-2 inhibitor etoricoxib attenuated hypoxic cancer milieu induced m2-polarization of macrophages and acquisition of pro-angiogenic and pro-invasive attributes Jain N.K., Baghel K.S.. Research Journal of Pharmacy and Technology.2019.
- A contemporary review on the critical role of nonsteroidal anti-inflammatory agents in colorectal cancer therapy Zia Sarabi P, Moradi M, Bagheri M, Khalili MR , Moradifard S, Jamialahmadi T, et al . Anti-Cancer Agents in Medicinal Chemistry-Anti-Cancer Agents).2024;24(8):559-70.
- Effect of dietary sesame oil with etoricoxib against 1, 2-dimethylhydrazine induced colon cancer in rats: Rajiv Gandhi University of Health Sciences (India) Patel HC . 2010.
- Esophageal adenocarcinoma. COX-2 Inhibitors Saukkonen K. 2024;:227.
- Structural tailoring of etoricoxib: A spectrochemical, medicinal and pharmacological study Akter B., Aishee S., Hridoy A., Pulok M.M.H., Islam M.A., Biswas A.. Chemical Physics Impact.2025.
- Theoretical investigation of the molecular structure and molecular docking of etoricoxib Sadasivam K., Salgado Moran G., Mendoza-Huizar L.H., Cardona Villada W., Gerli Candia L., Meneses-Olmedo L.M.. Journal of the Chilean Chemical Society.2020.
- Cancer cells dysregulate pi3k/akt/mtor pathway activation to ensure their survival and proliferation: Mimicking them is a smart strategy of gammaherpesviruses Cirone M.. Critical Reviews in Biochemistry and Molecular Biology.2021. CrossRef
- Pi3k/akt/mtor signaling pathway in cancer stem cells Ebrahimi M., Nourbakhsh E., Hazara A.Z., Mirzaei A., Shafieyari S., Salehi A.. Pathology-Research and Practice.2022.
- The pathogenic role of pi3k/akt pathway in cancer onset and drug resistance: An updated review Rascio F., Spadaccino F., Rocchetti M.T., Castellano G., Stallone G., Netti G.S.. Cancers.2021;13(16). CrossRef
- Angiogenic signaling pathways and anti-angiogenic therapy for cancer Liu Z.-L., Chen H.-H., Zheng L.-L., Sun L.-P., Shi L.. Signal transduction and targeted therapy.2023. CrossRef
- Targeting pi3k/akt/mtor signaling to overcome drug resistance in cancer Tufail M, Wan WD S, Jiang C C, Li N N. Chemico- Biological Interactions.2024;396. CrossRef
- Strategic advancements in targeting the pi3k/akt/mtor pathway for breast cancer therapy Garg P, Ramisetty S, Nair M, Kulkarni P, Horne D, Salgia R, et al . Biochemical Pharmacology.2025;236:116850.
- What hela cells aka immortal cells are and why they are important. An example of racism in medicine. Dr. Hakim Saboowala Saboowala HK . 2022.
- Hela cell culture: Immortal heritage of henrietta lacks Lyapun I., Andryukov B., Bynina M.. Molecular Genetics, Microbiology and Virology.2019.
- In vitro periodontal ligament cell expansion by co-culture method and formation of multi-layered periodontal ligament-derived cell sheets Safi I.N., Hussein B.M.A., Al-Shammari A.M.. Regenerative therapy.2019. CrossRef
- Cell line authentication: A necessity for reproducible biomedical research Souren N.Y., Fusenig N.E., Heck S., Dirks W.G., Capes‐Davis A., Bianchini F.. The EMBO Journal.2022. CrossRef
- Calculating half maximal inhibitory concentration (ic 50) values from glycomics microarray data using graphpad prism M Le Berre, JQ Gerlach, I Dziembała, M Kilcoyne. Glycan Microarrays: Methods and Protocols.2022. CrossRef
- The changing 50% inhibitory concentration (ic50) of cisplatin: A pilot study on the artifacts of the mtt assay and the precise measurement of density-dependent chemoresistance in ovarian cancer He Y., Zhu Q., Chen M., Huang Q., Wang W., Li Q.. Oncotarget.2016;7(43). CrossRef
- Chemical composition, evaluation of antiparasitary and cytotoxic activity of the essential oil of psidium brownianum mart ex. Dc Bezerra JN , Gomez MCV , Rolón M, Coronel C, Almeida-Bezerra JW , Fidelis KR , et al . Biocatalysis and Agricultural Biotechnology.2022;39:102247. CrossRef
- Plip: Fully automated protein–ligand interaction profiler Salentin S., Schreiber S., Haupt V.J., Adasme M.F., MJNar S. Nucleic Acids Res.2015. CrossRef
- Recent advances in molecular docking for the research and discovery of potential marine drugs Chen G., Seukep A.J., MJMd G. Mar Drugs.2020. CrossRef
- Charting the fragmented landscape of drug synergy Meyer C.T., Wooten D.J., Lopez C.F., Quaranta V.. Trends Pharmacol Sci.2020. CrossRef
- The combination index (ci< 1) as the definition of synergism and of synergy claims Chou T-CJS . Elsevier.2018;:49-50.
- Statistical analysis system, user's guide. Statistical. Version 9 Cary NJSIIU . 2012.
- Interaction effect of methotrexate and aspirin on mcf7 cell line proliferation: In vitro study Mahdi H.M., Wadee S.A.. Journal of Advanced Veterinary Research.2023.
- Qsar aided design of potent ret inhibitors using molecular docking, molecular dynamics simulation and binding free energy calculation Guo L.-Y., Xing X., Tong J.-B., Li P., Ren L., An C.-X.. Molecular Dynamics Simulation and Binding Free Energy Calculation.2024.
- Alpelisib plus fulvestrant in pik3ca-altered and pik3ca-wild-type estrogen receptor–positive advanced breast cancer: A phase 1b clinical trial Juric D., Janku F., Rodón J., Burris H.A., Mayer I.A., Schuler M.. JAMA oncology.2019. CrossRef
- Capivasertib Hitt E.M., Baker D.E.. Hospital Pharmacy.2024. CrossRef
- Phase ii clinical trial of everolimus in a pan-cancer cohort of patients with mtor pathway alterations Adib E., Klonowska K., Giannikou K., Do K.T., Pruitt-Thompson S., Bhushan K.. Clinical Cancer Research.2021. CrossRef
- Evaluation of lh, fsh, oestradiol, prolactin and tumour markers cea and ca-125 in sera of iraqi patients with endometrial cancer Dawood Y.J., Mahdi M.A., Jumaa A.H., Saad R., Khadim R.M.. Scripta Medica.2024;55(4).
- Jarad A. Diabetic wound healing enhancement by tadalafil. 2020. https://doi.org/10.31838/ijpr/2020.12.03.121. .
- The pi3k pathway in human disease Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T.. Cell.2017. CrossRef
- Targeting pi3k/akt/mtor signaling in cancer Porta C., Paglino C., Mosca A.. Frontiers in oncology.2014. CrossRef
- A phase ib study of alpelisib (byl719), a pi3kα-specific inhibitor, with letrozole in er+/her2− metastatic breast cancer Mayer I.A., Abramson V.G., Formisano L., Balko J.M., Estrada M.V., Sanders M.E.. Clinical cancer research.2017. CrossRef
- Development of pi3k inhibitors: Lessons learned from early clinical trials Rodon J., Dienstmann R., Serra V., Tabernero J.. Nature reviews Clinical oncology.2013. CrossRef
- Integrative hub of developmental and oncogenic signaling network Li T., Forbes M.E., Fuller G.N., Li J., Yang X., Igfbp2 Zhang W.. Oncogene.2020. CrossRef
- Glioma targeted therapy: Insight into future of molecular approaches Yang K., Wu Z., Zhang H., Zhang N., Wu W., Wang Z.. Molecular cancer.2022. CrossRef
- Phase i trial of the parp inhibitor olaparib and akt inhibitor capivasertib in patients with brca1/2-and non–brca1/2-mutant cancers Yap T.A., Kristeleit R., Michalarea V., Pettitt S.J., Lim J.S., Carreira S.. Cancer discovery.2020. CrossRef
- Cellular and molecular effects of the mtor inhibitor everolimus Saran U., Foti M., Dufour J.-F.. Clinical science.2015. CrossRef
- Phase 1 study of mtorc1/2 inhibitor sapanisertib (tak-228) in advanced solid tumours, with an expansion phase in renal, endometrial or bladder cancer Voss M.H., Gordon M.S., Mita M., Rini B., Makker V., Macarulla T.. British journal of cancer.2020. CrossRef
- Targeting the pi3k pathway in cancer: Are we making headway? Janku F., Yap T.A., Meric-Bernstam F.. Nature reviews Clinical oncology.2018. CrossRef
- P-selectin is a nanotherapeutic delivery target in the tumor microenvironment Shamay Y., Elkabets M., Li H., Shah J., Brook S., Wang F.. Science translational medicine.2016. CrossRef
- Β-catenin activation promotes immune escape and resistance to anti–pd-1 therapy in hepatocellular carcinoma Galarreta M Ruiz, E Bresnahan, P Molina-Sánchez, KE Lindblad, B Maier, D Sia. Cancer discovery.2019. CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2025
Author Details