LSP1 Gene rs3817198 Polymorphism and Breast Cancer Risk: A Systematic Review and Meta-analysis Study
Download
Abstract
In this meta-analysis, we tried to clear the relationship between breast cancer risk and LSP1 gene rs3817198T>C polymorphism. This meta-analysis was conducted according to PRISMA protocol. We searched PubMed/Medline, Web of sciences and EMBASE. All literature investigating the association of LSP1 gene rs3817198T>C and breast cancer risk were considered to include in the meta-analysis. We pooled ORs using both fixed and random-effect models. Egger’s test and funnel plot were used to evaluate Publication bias and small study effect. After evaluation and screening of citations, 14 publications were eligible for final analysis after applying of inclusion and exclusion criteria. Overall, 30,204 cases and 35,282 controls included in this meta-analysis. There was the significant association between LSP1 gene rs3817198T>C polymorphism and breast cancer only in homozygote genetic model (OR=1.14 [1.05-1.24]) and no association was found in heterozygotes (OR=1.03 [0.98-1.07]). The association was significant for popula ion-based studies and European & American & African population in both homozygote and heterozygote genetic model. There was no evidence of bias of literature and no small study effect. In conclusion, it seems that LSP1 gene rs3817198 polymorphism play its role in breast cancer incidence and other SNPs and environment are such triggers. Nevertheless, we recommend genome-wide association studies to evaluate the effect of SNPs in combination, not as single SNPs.
Introduction
Breast cancer accounts as the most frequent type of cancer among women and the incidence of breast cancer are rapidly increasing, particularly in developing countries. Breast cancer arises from the effect of both genetic and environmental factors [1]. Although, the role of single nucleotide polymorphisms (SNPs) in the incidence of breast cancer has been shown in many studies [2, 3, 4, 5], 30-40% of the familial and only 3-4% of the total number of breast cancer cases attributed to genetic component [6].
Leukocyte-specific protein 1 (LSP1) gene encoding an F-actin binding protein is located on 11p15.5. Studies investigating association LSP1 gene rs3817198T>C and breast cancer risk showed different results. However, there most of the studies showed no association between rs3817198 SNP and breast cancer [7, 8, 9], but there is evidence of increased [10, 11] or may be decreased risk of breast cancer [12]. In this meta-analysis, we tried to clear the relationship between breast cancer risk and LSP1 gene rs3817198T>C polymorphism.
Materials and Methods
Literature identification
This meta-analysis was conducted according to PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) protocol. In order to retrieve all studies related to LSP1 gene and breast cancer, a complete search of databases was performed. We searched PubMed/Medline, Web of sciences and EMBASE using phrases “LSP”, “LSP1”, “lymphocyte-specific protein”, “WP34” and “breast cancer”, “breast tumor”, and “breast neoplasm”. Detailed search strategy of databases is provided in supplementary Tabl 1. The additional search of the Reference list of final retrieved studies and related meta-analyses was carried out to include missed citations.
| First author | Year | Menopausal status | Heterozygote | Homozygote | |||
| Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | ||||
| Easton, D. F. | 2007 | Mix | 1.06 (1.02-1.11) | - | 1.17 (1.08-1.25) | - | |
| Garcia-Closas, M. (ER-positive) | 2008 | Mix | 1.13 (1.00–1.27) | 1.19 (1.10–1.28) | - | ||
| Garcia-Closas, M. (ER-negative) | 2008 | Mix | 1.01 (0.94–1.09) | 1.04 (0.99–1.09) | - | ||
| Barnholtz-Sloan, J. S. (African-American) | 2010 | Mix | 1.19 (0.93-1.52) | - | 0.47 (0.23-0.99) | - | |
| Barnholtz-Sloan, J. S. (White) | 2010 | Mix | 1.04 (0.87–1.25) | - | 1.27 (0.94-1.71) | - | |
| Gorodnova, T. V. | 2010 | Mix | - | 1.40 (0.86-2.21) | 2.40 (1.11-4.99) | ||
| Latif, A. | 2010 | Mix | 0.88 (0.67-1.16) | - | 0.98 (0.65-1.50) | - | |
| Tamimi, R. M. | 2010 | Mix | 1.09 (0.88-1.36) | - | 1.18 (0.80-1.74) | - | |
| Jiang, Y | 2011 | Mix | 0.89 (0.66-1.19) | - | 0.91 (0.24-3.46) | - | |
| Sueta, A. | 2011 | Mix | 0.97 (0.91- 1.03) | - | 1.03 (0.93-1.14) | - | |
| Butt, S | 2012 | Mix | 1.06 (0.87-1.30) | 1.09 (0.90-1.33) | 1.50 (1.08-2.09) | 1.53 (1.10-2.11) | |
| Shan, J. | 2012 | Mix | 1.15 (0.87-1.52) | - | 1.45 (0.90-2.34) | - | |
| Mizoo, T. | 2013 | Mix | 1.07 (0.77‐1.49) | 1.14 (0.85-1.55) | 1.63 (0.52‐5.66) | 2.04 (0.72-6.60) | |
| Pre | 1.00 (0.60-1.68) | 0.92 (0.58-1.48) | 3.29 (0.42-68.89) | 3.98 (0.58-78.39) | |||
| Post | 1.18 (0.75-1.86) | 1.30 (0.87-1.94) | 1.39 (0.32-6.31) | 1.65 (0.46-6.55) | |||
| Chen, Y. | 2016 | Mix | 0.61 (0.35-1.08) | - | 0.37 (0.08-1.66) | - | |
| Deng, Z. | 2016 | Mix | 0.99 (0.62-1.58) | - | 3.79 (1.25-11.50) | - | |
| Tan, T. | 2016 | Mix | - | 0.85 (0.63-1.14)* | - | 0.72 (0.19-2.28)* |
a) Year: year of publication; OR: odds ratio; CI: confidence interval
b) ORs calculated by authors via STATA software
Study selection
All literature investigating the association of LSP1 gene rs3817198T>C and breast cancer risk were considered to include in the meta-analysis. We excluded studies if they had the following criteria: (1) studies evaluating different polymorphisms of LSP1 gene; (2) studies had no control group; (3) studies providing no measure of association or information for calculation odds ratio (OR); (4) studies presenting only per-allele OR, not for four genetic models; (5) studies with breast cancer mortality and benign breast disease as main outcome; (6) case report and animal studies.
Data extraction
Screening of citations and extraction of the required information was carried out independently by to authors (ASM, MR). The following information was collected from included studies: name of first author, publication date, study design, source of controls (population-based or hospital-based), considered confounders in each models, genotyping methods, population ethnicity, total number of cases and controls, minor allele frequency and OR and their reported 95% confidence interval (CI) for homozygote and heterozygote inheritance models.
Study qualification
Four items, which might interfere with the results of the meta-analysis, including 1) source of a control group, 2) ethnicity, 3) menopausal status, and 4) sample size was considered to assess the quality of included studies. Quality of studies based on mentioned four items is presented in Supplementary Tabl 2.
| Subgroup | Studies No. | Homozygote | Studies No. | Heterozygote | |||||||||||
| Fixed model OR (95% CI) | Random model OR (95% CI) | Ph | I2 | T2 | Fixed model OR (95% CI) | Random model OR (95% CI) | Ph | I2 | T2 | ||||||
| Overall | 16 | 1.10 (1.07-1.14) | 1.14 (1.05-1.24) | 0 | 59.2 | 0.01 | 16 | 1.03 (1.00-1.06) | 1.03 (0.98-1.07) | 0.21 | 21.4 | 0 | |||
| Control source | |||||||||||||||
| Hospital based | 3 | 1.03 (0.93-1.14) | 1.01 (0.71-1.46) | 0.31 | 13.7 | 0.04 | 3 | 0.97 (0.91-1.03) | 0.95 (0.80-1.13) | 0.23 | 31.9 | 0.01 | |||
| Population based | 13 | 1.11 (1.07-1.15) | 1.17 (1.06-1.29) | 0 | 63.1 | 0 | 13 | 1.05 (1.02-1.09) | 1.05 (1.02-1.09) | 0.57 | 0 | 0 | |||
| Continent | |||||||||||||||
| Europe_america_africa | 8 | 1.18 (1.11-1.27) | 1.22 (1.03-1.44) | 0.07 | 47.4 | 0.02 | 8 | 1.06 (1.02-1.10) | 1.06 (1.02-1.10) | 0.74 | 0 | 0 | |||
| Asia | 6 | 1.04 (0.94-1.15) | 1.13 (0.70-1.81) | 0.16 | 37.1 | 0.12 | 6 | 0.96 (0.91-1.02) | 0.96 (0.91-1.02) | 0.56 | 0 | 0 |
OR: odds ratio; CI: confidence intervals; Ph: P values of Q-test for heterogeneity test; I2: I-square; T2: Tau-square
Statistics
Allelic frequency of controls calculated for studies separately. Calculated frequencies of the LSP1 gene rs3817198T>C genotype was evaluated for Hardy–Weinberg equilibrium using the X2 statistic. We pooled ORs using both fixed and the DerSimonian and Laird random-effects models. Heterogeneity across studies was assessed using Q and I2 statistics [13 14, ]. An I2 value above 75% at a significance level of < 0.1 was considered as presence of statistically significant heterogeneity [15]. Egger’s test and contour-enhanced funnel plot were used to evaluate Publication bias and small study effect [16]. The measure of association of all genetic inheritance models including (TT versus CC (homozygote) and TC vs. CC (heterozygote)) was assessed. Additional subgroup analysis was performed for ethnicity, and study setting. The ethnicity subgroup was defined based on the continent as Asians and Europeans & North Americans & Africans. Subgrouping included population-based (those reported a population-based case-control or nested case-control) and hospital-based (those reported a hospital-based case-control). All analyses were performed using Stata version 13 (Stata Corp LP, College Station, TX, USA).
Results
Selected literature
Through the primary search of the literature, 287 citations were identified. After evaluation and screening of citations, 29 publications considered for complete assessment of full-text. fourteen publications were eligible for final analysis after applying of inclusion and exclusion criteria [7, 8, 9, 10, 11, 12, 17, 18, 19, 20, 21, 22, 23, 24]. Figure one display the screening process(Fig. 1). Studies conducted by Barnholtz-Sloan (African-American & white) and Garcia-Closas (ER-positive & ER-negative) [12, 20] were considered as two separate studies each because provided ORs for two different populations. Table 1 shows adjusted and unadjusted OR for every single study. From fourteen, eleven had a population-based source of control. Five literatures were conducted in Europe, one in North America, six in Asia and one in Africa. Overall, 30,204 cases and 35,282 controls included in this meta-analysis. Detailed characteristic of included studies and reason of exclusion of others are provided in supplementary Table 3 and 4.
Figure 1 :flowchart diagram of selection process
Analysis
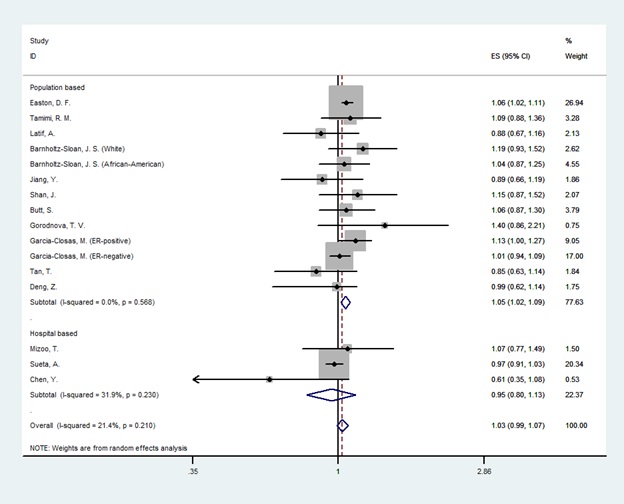
In the overall analysis, there was a significant association between LSP1 gene rs3817198T>C polymorphism and breast cancer only in homozygote genetic model (OR=1.14[1.05-1.24], (Fig. 2) and no association was found in heterozygotes (OR=1.03 [0.98-1.07], (Fig. 3). In the subgroup analysis by the source of controls and ethnicity, the significant association was found for population-based studies and European & American & African population in both homozygote and heterozygote genetic model. Nevertheless, results showed no relationship in Asians and hospital-based studies. The result of meta-analysis is presented in Table 2.
Figure 2 :Random effect model meta-analysis of LSP1 gene rs3817198 polymorphism for homozygote genetic models and risk of breast cancer. The effect size is odds ratio.
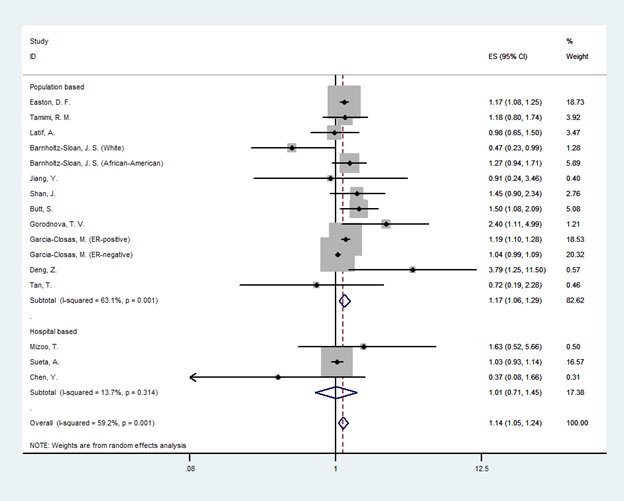
Figure 3 :Random Effect Model Meta-Analysis Of LSP1 Gene Rs3817198 Polymorphism For Heterozygote Genetic Models And Risk Of Breast Cancer. The Effect Size Is Odds Ratio.
Publication bias
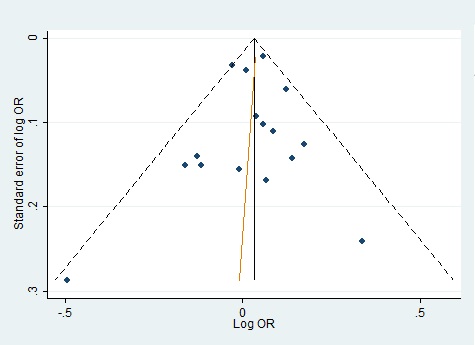
We evaluated publication bias by mean of Begg’s funnel plot and Egger’s test and there was no evidence of bias of literature and no small study effect (Fig. 4)(Fig. 5).
Figure 4 :Funnel Plot For Homozygote Genetic Model With Regression Line Corresponding To The Egger’s Regression Test For Funnel-Plot Asymmetry
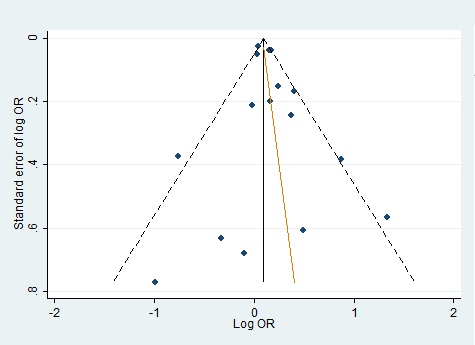
Figure 5 :Funnel plot for heterozygote genetic model with regression line corresponding to the Egger’s regression test for funnel-plot asymmetry.
Discussion
Genetic predisposition to breast cancer has been studied in many studies and some have proved the role of some SNPs in the incidence of breast cancer. LSP1 gene encodes an F-actin binding protein, which helps in neutrophil migration and chemotaxis [25]. The linkage between LSP1 gene rs3817198T>C mutation and breast cancer has been evaluated in several literatures, but there is no unanimity in the results. Some showed no association between LSP1 gene rs3817198 polymorphism and breast cancer [8] and some revealed increased risk of breast cancer coming with the polymorphism [10]. To bring together all the information, we conducted this meta-analysis to evaluate the association between LSP1 gene rs3817198 polymorphism and breast cancer in different designs and ethnicities and update the previous meta-analysis. Overall, we found that LSP1 gene rs3817198 polymorphism is associated with breast cancer only in homozygote genetic model. In subgroup analysis, the association was significant in population-based studies and European & American & African population in both homozygote and heterozygote genetic model.
The association between breast cancer and LSP1 gene rs3817198 polymorphism was only significant in homozygote genetic model, which was consistent with results of two previous meta-analyses [26, 27]. This result demonstrates that minor allele carries the risk of breast cancer. In subgroup analysis, we found the significant association for European & American & African population not Asian in both genetic models. The most recent meta-analysis [27] showed the same results but the earlier [26] documented the association only in homozygote genetic model. Although we categorized the population according to the continent and the two previous meta-analyses based on ethnicity, it makes sense that they considered European and Americans as Caucasian. Association between LSP1 gene rs3817198 polymorphism and breast cancer in European & American & African population, not Asian support the role of environment and also the possibility of the existence of concomitant SNPs. In addition, the association remained significant just in population-based studies. Nevertheless, this association was not seen in two mentioned studies. The diversity is probably because we included more studies and besides, we took nested case-control as population-based studies. As we know, the risk of bias is lower in population-based studies, so their results are more reliable. Significant association in population-based studies confirms the main results of this meta-analysis.
This paper faced some limitation. First, some studies did not provide adjusted ORs and we analyzed both adjusted and unadjusted ORs together. Second, due to lack of information, we were not able to perform subgroup analysis for menopausal status and other risk factors.
In conclusion, it seems that LSP1 gene rs3817198 polymorphism play its role in breast cancer incidence and other SNPs and environment are such triggers. Nevertheless, we recommend genome-wide association studies to evaluate the effect of SNPs in combination, not as single SNPs.
References
[1]. Xi J, Su Y, Beeghly Fadiel A, Lin Y, Su FX, Jia WH, et al. Association of physical activity and polymorphisms in FGFR2 and DNA methylation related genes with breast cancer risk. Cancer Epidemiol. 2014;38(6):708-14.
[2]. Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, et al. Novel breast cancer susceptibility locus at 9q31. 2: results of a genome-wide association study. Journal of the National Cancer Institute. 2011;103(5):425-35.
[3]. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353-61, 61e1-2.
[4]. Sanjari Moghaddam A, Nazarzadeh M, Sanjari Moghaddam H, Bidel Z, Keramatinia A, Darvish H, et al. XRCC1 Gene Polymorphisms and Breast Cancer Risk: A Systematic Review and Meta- Analysis Study. Asian Pac J Cancer Prev. 2016;17(S3):323-30.
[5]. Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504-7.
[6]. Lux MP, Fasching PA, Beckmann MW. Hereditary breast and ovarian cancer: review and future perspectives. J Mol Med (Berl). 2006;84(1):16-28.
[7]. Jiang Y, Han J, Liu J, Zhang G, Wang L, Liu F, et al. Risk of genome-wide association study newly identified genetic variants for breast cancer in Chinese women of Heilongjiang Province. Breast Cancer Res Treat. 2011;128(1):251-7.
[8]. Latif A, Hadfield KD, Roberts SA, Shenton A, Lalloo F, Black GC, et al. Breast cancer susceptibility variants alter risks in familial disease. J Med Genet. 2010;47(2):126-31.
[9]. Tamimi RM, Lagiou P, Czene K, Liu J, Ekbom A, Hsieh CC, et al. Birth weight, breast cancer susceptibility loci, and breast cancer risk. Cancer Causes Control. 2010;21(5):689-96.
[10]. Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087-93.
[11]. Gorodnova TV, Kuligina E, Yanus GA, Katanugina AS, Abysheva SN, Togo AV, et al. Distribution of FGFR2, TNRC9, MAP3K1, LSP1, and 8q24 alleles in genetically enriched breast cancer patients versus elderly tumor-free women. Cancer Genet Cytogenet. 2010;199(1):69-72.
[12]. Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31(8):1417-23.
[13]. Handoll HH. Systematic reviews on rehabilitation interventions. Arch Phys Med Rehabil. 2006;87(6):875.
[14]. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719-48.
[15]. DerSimonian R, Laird N. Meta-analysis in clinical trials Control Clin Trials 7.Find this article online. 1986;7(3):177–188.
[16]. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629-34.
[17]. Butt S, Harlid S, Borgquist S, Ivarsson M, Landberg G, Dillner J, et al. Genetic predisposition, parity, age at first childbirth and risk for breast cancer. BMC research notes. 2012;5:414.
[18]. Chen Y, Shi C, Guo Q. TNRC9 rs12443621 and FGFR2 rs2981582 polymorphisms and breast cancer risk. World J Surg Oncol. 2016;14(1):50.
[19]. Deng Z, Yang H, Liu Q, Wang Z, Feng T, Ouyang Y, et al. Identification of novel susceptibility markers for the risk of overall breast cancer as well as subtypes defined by hormone receptor status in the Chinese population. Journal of Human Genetics. 2016;61:1027–34.
[20]. Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4(4):e1000054.
[21]. Mizoo T, Taira N, Nishiyama K, Nogami T, Iwamoto T, Motoki T, et al. Effects of lifestyle and single nucleotide polymorphisms on breast cancer risk: a case-control study in Japanese women. BMC Cancer. 2013;13:565.
[22]. Shan J, Mahfoudh W, Dsouza SP, Hassen E, Bouaouina N, Abdelhak S, et al. Genome-Wide Association Studies (GWAS) breast cancer susceptibility loci in Arabs: susceptibility and prognostic implications in Tunisians. Breast Cancer Res Treat. 2012;135(3):715-24.
[23]. Sueta A, Ito H, Kawase T, Hirose K, Hosono S, Yatabe Y, et al. A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Research and Treatment. 2012;132(2):711-21.
[24]. Tan T, Zhang K, Chen W. Genetic variants of ESR1 and SGSM3 are associated with the susceptibility of breast cancer in the Chinese population. Breast Cancer. 2017;24(3):369-74.
[25]. Lanigan F, O'Connor D, Martin F, Gallagher WM. Molecular links between mammary gland development and breast cancer. Cellular and molecular life sciences: CMLS. 2007;64(24):3159-84.
[26]. Chen MB, Li C, Shen WX, Guo YJ, Shen W, Lu PH. Association of a LSP1 gene rs3817198T>C polymorphism with breast cancer risk: evidence from 33,920 cases and 35,671 controls. Mol Biol Rep. 2011;38(7):4687-95.
[27]. Tang J, Li H, Luo J, Mei H, Peng L, Li X. The LSP1 rs3817198 T > C polymorphism contributes to increased breast cancer risk: a meta-analysis of twelve studies. Oncotarget. 2016;7(39):63960-7.
License
Copyright
© ,
Author Details