The Role of the Cadherin (CDH) Gene Family in the Carcinogenic Processes of Ovarian Cancer: A Comprehensive Bioinformatics Analysis
Download
Abstract
Background: The persistent challenge of ovarian cancer as a major driver of cancer mortality in the female population stems largely from its tendency toward late-stage identification and frequent disease relapse. The cadherin (CDH) gene family, crucial for cell-cell adhesion, plays complex roles in cancer progression.
Objective: Bioinformatics analysis of the CDH gene family in ovarian cancer. Using multiple public databases.
Methodology: Transcriptome analysis of cadherin (CDH) gene family in ovarian cancer was performed using Gene Expression Profiling Interactive Analysis 2 (GEPIA2). Prognostic value of differentially expressed CDH genes was assessed using Kaplan-Meier plotter Overall Survival (OS) . Protein-level validation was performed using Human Protein Atlas (HPA) portal which provides immunohistochemistry (IHC). By using GSCALite web server, the assessment of immune cell infiltration was conducted to explore correlations between cadherin expression and tumor immune microenvironment and drug sensitivity analysis was performed to evaluate candidate CDH genes as therapeutic response predictors.
Results: Our findings revealed significant differential expression of several CDH genes: CDH1 and CDH4 were downregulated while CDH2, CDH6, CDH11, and CDH23 were upregulated in ovarian cancer tissues. Survival analysis identified CDH6, CDH11, and CDH23 as adverse prognostic markers correlating with poorer overall and progression-free survival, while high CDH2 and CDH4 expression associated with improved survival. Genetic alteration analysis revealed diverse genomic changes across the CDH family, with protein expression data largely corroborating transcriptomic findings. Novel associations between CDH expression and drug sensitivity emerged as potential predictive biomarkers. CDH1 and CDH11 expression correlated with Paclitaxel and Dasatinib resistance, respectively, while CDH2 and CDH6 expression indicated sensitivity to PI3K and Src kinase inhibitors.
Conclusion: This study provides comprehensive molecular characterization of CDH family roles in ovarian cancer progression, prognosis, drug response, and immune regulation, establishing specific CDH members as potential diagnostic and therapeutic targets for ovarian cancer.
Introduction
Ovarian cancer is the deadliest gynecological malignancy, primarily due to asymptomatic early stages and late-stage detection when metastasis has occurred [1]. Advanced-stage ovarian cancer survival remains approximately 30%, unchanged despite sophisticated surgical and chemotherapeutic advances [2, 3]. This underscores the critical need for novel molecular biomarkers enabling early diagnosis, improved prognosis, and precision medicine strategies [4].
Cell-cell adhesion, dynamically regulated by the cadherin (CDH) family of calcium-dependent transmembrane glycoproteins, plays crucial roles in ovarian cancer progression and metastasis [5]. Cadherins maintain cellular polarity and tissue architecture, and their dysfunction characterizes cancer, particularly during epithelial-mesenchymal transition (EMT) [6]. During EMT, epithelial cells lose intercellular adhesion mediated by E-cadherin (CDH1) and acquire mesenchymal phenotypes with increased motility and invasiveness. This “cadherin switching” E-cadherin downregulation with N-cadherin (CDH2) upregulation enables tumor cell dissemination and distant metastasis [7, 8]. The CDH family comprises numerous members with diverse tissue-specific expression and functions [9]. While CDH1 and CDH2 roles are well-characterized across cancers, contributions of less-studied CDH family members to ovarian carcinogenesis remain largely unexplored [10]. Individual studies have implicated specific cadherins like P-cadherin (CDH3) and Cadherin-6 (CDH6) in ovarian cancer cell migration and invasion [11, 12], but comprehensive pan-family analysis is lacking. High-throughput technologies and major public cancer genomics collections, including The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects, now enable unprecedented comprehensive bioinformatics analyses [13, 14]. Integrating transcriptomic, genomic, and clinical data allows systematic exploration of CDH gene family expression patterns, prognostic value, and roles in carcinogenic pathways.
This study presents the first comprehensive bioinformatics analysis of the CDH gene family in ovarian cancer. Using multiple public databases, we identify differentially expressed CDH genes, assess their impact on patient survival, and examine associations with clinical characteristics, aiming to discover novel CDH-derived biomarkers for improved prognostic stratification and therapeutic management of ovarian cancer patients.
Materials and Methods
Differential Expression of CDH Family Members
Transcriptome analysis of cadherin (CDH) gene family in ovarian cancer was performed using Gene Expression Profiling Interactive Analysis 2 (GEPIA2) web server (http://gepia2.cancer-pku.cn), which combines RNA-Seq data from 9,736 tumor and 8,587 normal tissue samples from TCGA and GTEx projects [15]. Expression profiles of CDH family members were compared between ovarian cancer and normal tissues using independent Student’s t-test, with statistical significance set at (p< 0.05).
Prognostic Survival Analysis
Prognostic value of differentially expressed CDH genes was assessed using Kaplan-Meier plotter (http:// kmplot.com/analysis/) for ovarian cancer cohorts [16]. Overall Survival (OS) and Progression-Free Survival (PFS) were analyzed as primary endpoints. Patients were categorized into high- and low-expression groups using auto-selected optimal cut-off points. Statistical significance was determined using log-rank p-values, with hazard ratios (HRs) and 95% confidence intervals (CIs) calculated to measure effect sizes.
Genomic Alteration and Interaction Network Analysis
The genomic characterization of prognostically relevant CDH genes was conducted through the cBioPortal platform (http://www.cbioportal.org) [17, 18]. The Ovarian Serous Cystadenocarcinoma (TCGA, Firehose Legacy) dataset was interrogated to determine genomic alteration frequencies and types (mutations, copy number variations) for CDH1, CDH2, CDH4, CDH6, CDH11, and CDH23.
Transcriptional co-expression patterns were assessed using pairwise correlation matrix analysis of mRNA expression data. Protein-protein interaction (PPI) networks were generated using STRING database (v12.0) to explore functional relationships [19].
Immunohistochemical Validation of Protein Expression
Protein-level validation was performed using Human Protein Atlas (HPA) portal (http://www.proteinatlas.org) [20], which provides immunohistochemistry (IHC) data for 44 normal tissues and 20 cancer types. High-resolution IHC images were analyzed to compare cadherin protein expression patterns between serous ovarian carcinoma and normal ovarian samples. Staining intensity was graded using HPA’s four-level system: strong, moderate, weak, and negative, enabling assessment of target protein abundance in the tumor microenvironment.
Analysis of Gene-Gene Interactions
Functional network analysis was performed using GeneMANIA web server (http://www.genemania.org) to explore CDH gene family interactions and biological functionality [21]. The platform integrates genomic and proteomic datasets to construct functional association networks. All known human CDH family members were analyzed to generate evidence-based networks incorporating co-expression, physical interactions, shared protein domains, and co-localization data, enabling visualization of the CDH family functional landscape and identification of prominent shared pathways.
Pathway and Gene Ontology Enrichment Analysis
Functional enrichment analysis was performed to identify biological processes associated with the six candidate CDH genes. The 1,000 most co-expressed genes were identified using GEPIA2’s “Similar Genes” module from TCGA ovarian cancer data. Combined candidate and co-expressed gene lists were analyzed using Enrichr web server (https://maayanlab.cloud/Enrichr) [22] for over-representation analysis across Gene Ontology categories (Biological Process, Cellular Component, Molecular Function) and Reactome pathways (2024). The top ten most significant terms (p< 0.05) from each category were selected for interpretation.
Analysis of Tumor-Infiltrating Immune Cells
The assessment of immune cell infiltration was conducted through the GSCALite web server [23], to explore correlations between cadherin expression and tumor immune microenvironment. The “Immune Infiltration” module determined correlations between candidate CDH gene mRNA expression and abundance of 24 immune cell types in ovarian cancer cohorts, calculated using the ImmuCellAI algorithm. Correlations were measured via rank correlation coefficients between gene expression and immune cell abundance. Significant correlations were defined as |p|> 0.2 with false discovery rate (FDR) < 0.05.
Prediction of Therapeutic Drug Response
Drug sensitivity analysis was performed using GSCALite web server [23], to evaluate candidate CDH genes as therapeutic response predictors. The platform integrates drug sensitivity data from Genomics of Drug Sensitivity in Cancer (GDSC) database, containing IC50 values for hundreds of small-molecule inhibitors tested in cancer cell lines, with mRNA expression data. Analysis focused on ovarian cancer cell lines to determine correlations between CDH gene expression and drug sensitivity. Spearman rank correlation assessed relationships between gene expression and drug IC50 values. Positive correlations indicated drug resistance (higher expression = higher IC50), while negative correlations suggested drug sensitivity (higher expression = lower IC50). Significant drug-gene pairs were defined as |p| > 0.2 with FDR < 0.05.
Results
Differential Expression of CDH Family Members in Ovarian Cancer
To investigate cadherin (CDH) gene family roles in ovarian carcinogenesis, we analyzed differential expression between ovarian cancer and normal tissues using TCGA and GTEx data. Box plot analysis of 23 CDH members revealed significant dysregulation patterns (Figure 1).
Figure 1. Differential Expression of Cadherin (CDH) Family Members in Ovarian Cancer (OV) Tissues Compared to Normal Tissues. Box plots show the transcript levels (Log2(TPM+1)) for each gene. The red boxes represent tumor samples (T) and the grey boxes represent normal samples (N). Red asterisks (*) indicate a statistically significant difference (p< 0.05).
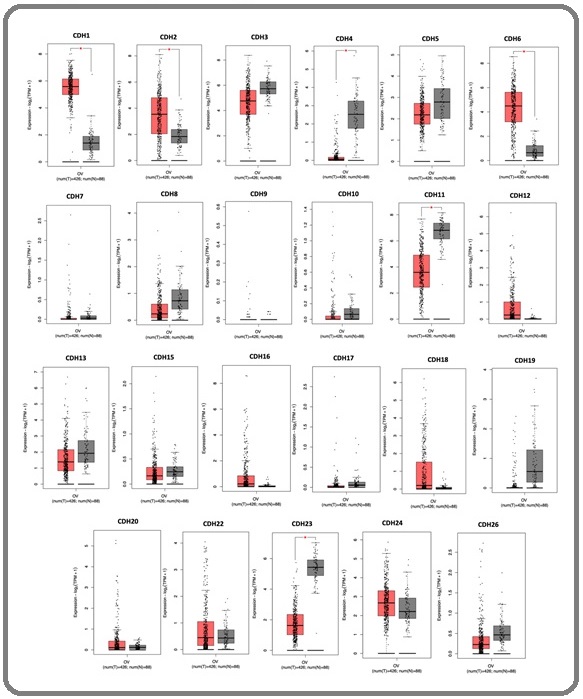
Three genes showed significant upregulation in tumors: CDH1, CDH2, and CDH6 (p< 0.05), with CDH1 and CDH6 demonstrating the most pronounced overexpression, suggesting tumor-promoting roles. Conversely, CDH4, CDH11, and CDH23 were significantly downregulated (p< 0.05), indicating potential tumor suppressor functions. CDH3 showed upregulation trends, while CDH7, CDH8, CDH9, and CDH10 exhibited no significant changes. Based on these findings, six genes with the most significant differential expression-CDH1, CDH2, CDH4, CDH6, CDH11, and CDH23-were selected for subsequent prognostic and functional analyses.
Prognostic Significance of Differentially Expressed CDH Genes in Ovarian Cancer
Survival analysis using Kaplan-Meier plotter assessed the prognostic value of six differentially expressed CDH genes (Table 1).
| Gene | Overall Survival (OS) | Progression-Free Survival (PFS) | ||
| HR (95% CI) | p value | HR (95% CI) | p value | |
| CDH1 | 0.87 (0.75–1.01) | 0.066 | 1.12 (0.98–1.3) | 0.1 |
| CDH2 | 0.81 (0.69–0.94) | 0.0054 | 0.82 (0.7–0.95) | 0.009 |
| CDH4 | 0.82 (0.71–0.95) | 0.0062 | 0.9 (0.78–1.03) | 0.11 |
| CDH6 | 1.24 (1.07–1.44) | 0.0047 | 1.38 (1.21–1.58) | 1.2e-06 |
| CDH11 | 1.38 (1.12–1.69) | 0.0022 | 1.4 (1.14–1.72) | 0.0013 |
| CDH23 | 1.34 (1.07–1.68) | 0.01 | 1.61 (1.31–1.98) | 5e-06 |
High expression of CDH6, CDH11, and CDH23 significantly correlated with poor prognosis, showing reduced Overall Survival (HR=1.24, p=0.0047; HR=1.38, p=0.0022; HR=1.34, p=0.01) and Progression- Free Survival (HR=1.38, p=1.2e-06; HR=1.4, p=0.0013;HR=1.61, p=5e-06), suggesting oncogenic roles in Table1 and visualized in Figure 2.
Figure 2. Kaplan-Meier Survival Curves for Selected CDH Genes in Ovarian Cancer. The plots illustrate the association between high (red line) and low (black line) gene expression with (A) Overall Survival (OS) and (B) Progression-Free Survival (PFS).

Conversely, elevated CDH2 and CDH4 expression associated with favorable Overall Survival (HR=0.81, p=0.0054; HR=0.82, p=0.0062), with CDH2 also improving PFS (HR=0.82, p=0.009), indicating tumor-suppressive functions. CDH1 showed no significant association with OS (p=0.066) or PFS (p=0.1), despite trending toward poorer outcomes. These findings reveal heterogeneous prognostic roles within the CDH family, identifying CDH6, CDH11, and CDH23 as poor prognostic indicators while highlighting protective roles for CDH2 and CDH4.
Genomic Alterations and Interaction Network of Prognostic CDH Genes
Genomic alteration analysis using cBioPortal revealed that the six CDH genes were altered in 30% of serous ovarian cancer samples, with amplification as the predominant alteration type (Figure 3A).
Figure 3. Genomic Alterations, Co-expression Patterns, and Protein-protein Interaction Network of Prognostic CDH Genes in Ovarian Cancer. (A) OncoPrint summary from cBioPortal showing the frequency and type of genomic alterations for the six selected CDH genes. (B) Correlation matrix illustrating the pairwise transcriptional co-expression relationships among the genes. Red circles indicate positive correlation, and blue circles indicate negative correlation; the size and intensity of the circle correspond to the strength of the correlation.(C) Protein-protein interaction (PPI) network from the STRING database showing the predicted functional associations among the six cadherin proteins.

CDH4 and CDH6 showed highest alteration frequencies (7% each), followed by CDH2 and CDH1 (6% each), CDH23 (4%), and CDH11 (3%). CDH2 exhibited amplifications and truncating mutations, while CDH1 showed deep deletions, indicating copy number variations as key dysregulation mechanisms. Transcriptional co-expression analysis (Figure 3B) revealed strong negative correlation between CDH23 and other genes, particularly CDH1 and CDH2, suggesting antagonistic regulation. CDH6 and CDH11 showed moderate positive correlation, indicating potential co-regulation. Protein-protein interaction network analysis using STRING database (Figure 3C) demonstrated high interconnectivity among the six cadherins, suggesting they function as a complex module collectively influencing cell adhesion and carcinogenic processes rather than operating independently.
Validation of CDH Protein Expression in Ovarian Cancer Tissues
Immunohistochemistry validation using Human Protein Atlas data largely corroborated transcriptomic findings (Figure 4).
Figure 4. Immunohistochemical Analysis of Candidate CDH Protein Expression in Ovarian Cancer and Normal Ovarian Tissues. Representative images from the Human Protein Atlas show the protein expression and localization of CDH1, CDH2, CDH6, CDH11, and CDH23. For each gene, images of normal ovary tissue and serous cystadenocarcinoma are shown, along with a magnified view and a summary bar chart of staining intensities across multiple samples.

CDH1 protein showed medium-to- strong expression in serous cystadenocarcinoma samples but was absent in normal ovarian tissue, consistent with mRNA upregulation. CDH6 and CDH23 exhibited low-to- moderate tumor expression while being absent in normal tissue, supporting their tumorigenic roles. However, CDH2 and CDH11 protein patterns inversely correlated with mRNA levels. CDH2 showed negative tumor staining despite transcript upregulation, though it was strongly expressed in normal ovarian stroma. CDH11 was strongly expressed in normal tissue but negative in tumor samples, despite mRNA upregulation. These discrepancies suggest complex post-transcriptional regulatory mechanisms modulating final protein levels. CDH4 protein data was unavailable due to pending HPA database annotations.
Overall, IHC analysis confirms CDH1, CDH6, and CDH23 protein presence in ovarian tumors, providing additional evidence for their carcinogenic involvement.
Analysis of Gene-Gene Interactions
We used the GeneMANIA database to draw the functional relationships of the CDH gene family. The resulting network showed that there were substantial connections among the CDH genes and the rest of similar proteins like desmosomal cadherins (Figure 5).
Figure 5. The Human CDH Gene Family is Studied by Functional Interaction Network and Enrichment analysis. The network obtained via GeneMANIA demonstrates the wide and cross-functional connections of cadherin (CDH) and desmoglein (DSG/DSC) family’s members. The various colors of the connecting lines (edges) vary with the nature of evidence supporting the interactions made, such as co-expression, physical interaction, and shared protein domains. The pie chart in each node (gene) would indicate a percent composition that indicated its functions to the significantly enriched biological processes. Based on the analysis of the network, one can see that its enrichment comprises mostly functions associated with cell-cell junction organization, cell-cell adhesion as well as keratinocyte differentiation.

The genes were found to be primarily enriched in biological processes involved in the organization of cell-cell junctions, cell-cell adhesion, and cell keratinization as well as differentiation of keratinocytes with an enrichment analysis carried out on this network.
Functional Enrichment Analysis of Candidate CDH Genes
Functional enrichment analysis using Enrichr database examined the six candidate CDH genes and their 1000 co-expressed partners (Table S1) to identify significantly enriched pathways (Figure 6).
Figure 6. Functional Enrichment Analysis of Candidate CDH Genes and their Co-expressed Network in Ovarian Cancer. The bar charts display the top ten most significantly enriched terms for the six candidate CDH genes and their primary co-expressed partners, based on data retrieved from the Enrichr database. The analysis is categorized into four functional groups: Biological Process (BP), Cellular Component (CC), Molecular Function (MF), and Reactome pathways. The length of the bars corresponds to the significance of the enrichment (p< 0.05).

Gene ontology analysis revealed primary enrichment in biological processes related to tumor microenvironment remodeling, including “Extracellular Matrix Organization” (GO:0030198), “Extracellular Structure Organization” (GO:0043062), and “Regulation of Cell Migration” (GO:0030334), indicating roles beyond cell adhesion in tissue architecture modulation critical for invasion and metastasis. Cellular Component analysis confirmed protein localization to adhesive structures, with top terms being “Collagen- Containing Extracellular Matrix” (GO:0062023), “Actin Cytoskeleton” (GO:0015629), and “Cell-Substrate Junction” (GO:0030055). Molecular Function analysis indicated involvement in growth factor signaling and enzyme activity, including “Platelet-Derived Growth Factor Binding” (GO:0048407), “Cell-Cell Adhesion Mediator Activity” (GO:0098632), and “Metalloendopeptidase Activity” (GO:0004222). Reactome pathway analysis confirmed enrichment in “Extracellular Matrix Organization,” “Collagen Degradation,” and “Integrin Cell Surface Interactions.” These findings establish the candidate CDH genes as key components regulating cell adhesion, migration, and extracellular matrix remodeling canonical cancer progression features.
Association of CDH Gene Expression with Immune Cell Infiltration
Immune infiltration analysis using TISIDB database revealed significant correlations between CDH1, CDH2, CDH11, and CDH23 expression and specific immune cell populations in ovarian cancer (Table 2).
| Gene symbol | Immune cell type | Spearman correlation coefficient | P-value | FDR |
| CDH1 | Exhausted | -2.03E-01 | 3.37E-04 | 1.68E-03 |
| CDH11 | Bcell | -3.47E-01 | 3.58E-10 | 8.64E-08 |
| CDH11 | CD4_T | 2.16E-01 | 1.32E-04 | 7.69E-04 |
| CDH11 | Central_memory | 3.20E-01 | 8.62E-09 | 1.45E-07 |
| CDH11 | InfiltrationScore | 2.42E-01 | 1.68E-05 | 1.57E-04 |
| CDH11 | Macrophage | 2.60E-01 | 3.68E-06 | 2.97E-05 |
| CDH11 | Monocyte | 3.60E-01 | 6.59E-11 | 1.82E-08 |
| CDH11 | Neutrophil | -3.15E-01 | 1.60E-08 | 1.87E-07 |
| CDH11 | Tfh | 2.60E-01 | 3.60E-06 | 2.95E-05 |
| CDH11 | Th17 | -2.40E-01 | 2.04E-05 | 2.45E-04 |
| CDH11 | Th2 | 2.84E-01 | 3.67E-07 | 1.58E-05 |
| CDH11 | iTreg | 2.44E-01 | 1.42E-05 | 1.62E-04 |
| CDH11 | nTreg | 2.21E-01 | 8.93E-05 | 2.58E-03 |
| CDH2 | CD8_naive | 3.28E-01 | 3.69E-09 | 4.65E-08 |
| CDH2 | Cytotoxic | -2.23E-01 | 7.43E-05 | 4.74E-04 |
| CDH2 | Effector_memory | -2.38E-01 | 2.42E-05 | 2.34E-04 |
| CDH2 | Exhausted | -2.09E-01 | 2.11E-04 | 1.13E-03 |
| CDH2 | Macrophage | -2.61E-01 | 3.32E-06 | 2.72E-05 |
| CDH2 | Neutrophil | 2.45E-01 | 1.36E-05 | 8.30E-05 |
| CDH2 | Tfh | -2.34E-01 | 3.33E-05 | 2.13E-04 |
| CDH2 | iTreg | -2.09E-01 | 2.15E-04 | 1.68E-03 |
| CDH23 | Effector_memory | -2.41E-01 | 1.90E-05 | 1.92E-04 |
CDH11 showed the most extensive associations, positively correlating with pro-tumoral cells (macrophages, monocytes, central memory T cells, Tfh cells, Th2 cells, iTreg, nTreg) while negatively correlating with B cells, neutrophils, and Th17 cells, suggesting a role in creating an immunosuppressive microenvironment. Conversely, CDH2 expression associated with anti-tumoral immunity, showing positive correlation with CD8+ naive T cells but negative correlation with exhausted T cells, cytotoxic cells, effector memory T cells, macrophages, Tfh cells, and iTregs, indicating links to effective anti-tumor responses. CDH1 expression negatively correlated with exhausted T cells, while CDH23 negatively correlated with effector memory T cells (detailed results in Table S2). These findings highlight distinct immunomodulatory roles of CDH family members and suggest their potential as biomarkers for ovarian tumor immune status.
CDH Gene Expression as a Predictor of Drug Sensitivity
Drug sensitivity analysis using GSCA database correlated CDH gene expression with IC50 values in ovarian cancer cell lines to explore therapeutic utility (Table S3). High CDH1 expression associated with Paclitaxel resistance, while elevated CDH11 correlated with Dasatinib resistance. Conversely, high CDH2 expression predicted sensitivity to PI3K inhibitors Pictilisib and Taselisib, and elevated CDH6 expression indicated sensitivity to Src kinase inhibitor Saracatinib. These findings suggest CDH family members as potential biomarkers for treatment response prediction. CDH1 and CDH11 overexpression may identify patients unlikely to benefit from Paclitaxel and Dasatinib, respectively, while CDH2 and CDH6 overexpression may indicate candidates for PI3K or Src kinase inhibitor therapy.
Detailed correlations are provided in (Table S4).
Discussion
This multifactorial bioinformatics study aimed to explain the complex functions of the Cadherin (CDH) gene family and its complicated role in the development of ovarian cancer, utilizing the large-scale genomic and transcriptomic data. The results of our study bring new knowledge about the expression differences, prognostic values, genetic mutations, protein expression patterns, functional pathways, sensitivity to drugs, and immune microenvironment relation of the most important members of the CDH family in the context of ovarian cancer. This combined evidence supports the concept that cadherins constitute a complex and, in many cases, conflicting system in ovarian cancer development, which makes cadherins promising diagnostic, prognostic, and treatment targets.
Our analysis revealed significant differential expression of several CDH family members, with CDH4, CDH11, and CDH23 downregulated while CDH1, CDH2, and CDH6 were upregulated in ovarian cancer tissues. These findings align with established cadherin roles in cancer progression. Our findings supports the trend of E-cadherin (CDH1) upregulation in the early stages of epithelial ovarian carcinoma [24, 25]; an observation that is aligned with many other cancers where CDH1 is upregulated early [26]. The increase helps tumor cells maintain epithelial characteristics and form compact cell clusters. With tumor progression, however, CDH1 is downregulated as a hallmark of epithelial-mesenchymal transition (EMT), which is also critical for cancer invasion and metastasis [27]. Conversely, N-cadherin (CDH2) and CDH6 upregulation promotes cancer migration and invasiveness [28, 29].
Survival analysis demonstrated that high expression of CDH6, CDH11, and CDH23 significantly correlated with poorer overall survival and progression-free survival, reinforcing their potential as adverse prognostic biomarkers. Interestingly, higher CDH2 and CDH4 expression associated with improved survival outcomes. While N-cadherin’s oncogenic role is recognized, its association with improved survival warrants investigation, possibly reflecting the complex biological context- dependent roles or compensatory mechanisms [30]. Several explanations may account for this observation. CDH2 and CDH4 exhibit distinct stage-dependent functions: N-cadherin (CDH2) may preserve cell cohesion in early-stage tumors but promotes invasion in advanced disease [31], while R-cadherin (CDH4) demonstrates the opposite pattern, increasing in early stages and subsequently decreasing in advanced tumors with metastatic potential [32, 33]. The tumor microenvironment also influences CDH2 function, with immune cell infiltration potentially modifying the relationship between N-cadherin expression, disease progression, and patient outcomes [34,35].
Investigation of the genetic landscape revealed various CDH family alterations including mutations, amplifications, and deletions, which may directly affect transcriptional regulation and protein function. These genomic changes complement our transcriptomic findings, with upregulation potentially resulting from amplification and downregulation from deletions or inactivating mutations, particularly in tumor suppressor cadherins like CDH1 [36]. Protein expression validation using Human Protein Atlas confirmed the presence of CDH6 and CDH23 proteins in ovarian tumors, supporting their significance in ovarian carcinogenesis based on upregulation and poor outcome correlations.
Discrepancies between messenger RNA (mRNA) abundance and corresponding protein levels have been observed for cadherin genes CDH2 (N-cadherin) and CDH11 (Cadherin-11) in certain cancer tissues. This phenomenon represents a documented complexity in cancer biology, highlighting the critical influence of post-transcriptional regulatory mechanisms [37]. Several molecular processes may account for this discordance. Post-transcriptional gene silencing by microRNAs (miRNAs) represents one key mechanism. The miR-200 family, for example, indirectly targets N-cadherin by suppressing ZEB1/2, transcriptional repressors of E-cadherin [38]. Accelerated protein degradation by proteolytic enzymes such as matrix metalloproteinases (MMPs) within the tumor microenvironment provides another explanation [39]. Additionally, altered subcellular localization of cadherin proteins may contribute to this observational bias, as cadherins can be internalized from the cell surface or translocated away from cell-cell junctions [40].
Gene-gene interaction networks revealed extensive connections among the six candidate CDH genes and their co-expression partners, indicating these cadherins function as an integrated complex rather than independently. This aligns with cell adhesion molecules frequently organizing complexes and signaling centers to control cellular functions [41]. Functional enrichment analysis identified significant enrichment in biological processes related to extracellular matrix organization, extracellular structure organization, and cell migration regulation. These findings align with established cadherin functions in cell-cell contact and tumor microenvironment regulation [42]. Cellular component analysis confirmed localization to adhesive structures including actin cytoskeleton, cell-substrate junctions, and collagen-containing extracellular matrix. Molecular function analysis revealed involvement in platelet-derived growth factor binding, cell-cell adhesion mediator activity, and metalloendopeptidase activity, indicating CDH family members integrate into complex signaling pathways beyond structural roles.
Drug sensitivity analysis revealed substantial correlations between CDH gene expression and ovarian cancer cell line sensitivity to various small-molecule inhibitors, suggesting CDH expression as potential therapeutic biomarkers. High CDH1 expression correlated with Paclitaxel resistance, while elevated CDH11 associated with Dasatinib resistance. Conversely, high CDH2 expression predicted sensitivity to PI3K inhibitors Pictilisib and Taselisib, and elevated CDH6 indicated Saracatinib sensitivity. These findings justify future mechanistic research and may inform personalized treatment strategies.
CDH gene expression showed significant correlations with immune cell infiltration patterns. CDH11 demonstrated the most extensive associations, positively correlating with immunosuppressive cells (macrophages, monocytes, regulatory T cells) while negatively correlating with B cells, neutrophils, and Th17 cells, suggesting a role in creating immunosuppressive microenvironments. Conversely, CDH2 expression associated with favorable immune landscapes, correlating positively with CD8+ naive T cells and negatively with exhausted T cells, indicating potential for enhanced anti-tumor immune responses.
This analysis has key limitations including reliance on observational data from public datasets, semi-quantitative protein expression data, and lack of experimental validation. Future research should focus on experimental validation of prognostic biomarkers in independent cohorts, mechanistic investigation of CDH-drug sensitivity relationships, and deeper exploration of CDH roles in tumor immune microenvironments. Integration of multi-omics data with single-cell technologies could provide comprehensive insights into CDH family functions in ovarian cancer progression.
In conclusions, this bioinformatics analysis reveals the complex roles of the CDH gene family in ovarian cancer. We identified CDH6, CDH11, and CDH23 as adverse prognostic markers, while CDH2 and CDH4 were associated with improved survival. Beyond their traditional cell adhesion functions, CDH genes demonstrated significant associations with drug sensitivity patterns and immune cell infiltration, suggesting their potential as predictive biomarkers for personalized therapy and immunotherapy response. The CDH family represents a multifaceted player in ovarian cancer biology with promise as diagnostic, prognostic, and therapeutic targets. While our findings are hypothesis-generating and include some contradictory observations, they reflect the inherent complexity of cancer biology, where malignancy involves multi-level molecular interactions. These apparent contradictions underscore the importance of multi-platform analysis and the necessity of using diverse bioinformatics tools for comprehensive validation. Our study provides a foundation for future experimental validation, emphasizing the critical need for laboratory studies and clinical translation. Such investigations will deepen our understanding of the cadherin family’s role in ovarian cancer, resolve the observed contradictions, and ultimately improve patient outcomes. Despite methodological challenges, this work represents a valuable contribution to the scientific literature by highlighting the multifaceted nature of cadherin function in cancer biology.
Acknowledgments
None.
Conflict of Interests
The authors declared that no conflict of interest exists.
References
- Epithelial ovarian cancer Lheureux S, Gourley C, Vergote I, Oza AM . The Lancet.2019;393(10177). CrossRef
- Epidemiology of ovarian cancer: a review Brett M. R, Jennifer B. P, Thomas A. S, Brett M. R, Jennifer B. P, Thomas A. S. Cancer Biology & Medicine.2017;14(1). CrossRef
- Cancer statistics, 2022 Siegel RL , Miller KD , Fuchs HE , Jemal A. CA: A Cancer Journal for Clinicians.2022;72(1). CrossRef
- Molecular Biomarkers for the Early Detection of Ovarian Cancer Zhang R, Siu MKY , Ngan HYS , Chan KKL . International Journal of Molecular Sciences.2022;23(19). CrossRef
- The cell-cell adhesion molecule E-cadherin Van Roy F., Berx G.. Cellular and Molecular Life Sciences.2008;65(23). CrossRef
- Molecular mechanisms of epithelial–mesenchymal transition Lamouille S, Xu J, Derynck R. Nature Reviews Molecular Cell Biology.2014;15(3). CrossRef
- The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges Loh C, Chai J, Tang T, Wong W, Sethi G, Shanmugam M, Chong P, Looi C. Cells.2019;8(10). CrossRef
- Dual role of E-cadherin in cancer cells Rubtsova SN , Zhitnyak IY , Gloushankova NA . Tissue Barriers.2022;10(4). CrossRef
- Molecular evolution of the cadherin superfamily Hulpiau P, Van Roy F. The International Journal of Biochemistry & Cell Biology.2009;41(2). CrossRef
- Comprehensive analysis of prognostic significance of cadherin (CDH) gene family in breast cancer Ku S, Liu H, Su C, Yeh I, Yen M, Anuraga G, Ta HDK , et al . Aging.2022. CrossRef
- P-Cadherin Promotes Ovarian Cancer Dissemination Through Tumor Cell Aggregation and Tumor–Peritoneum Interactions Usui A, Ko SY , Barengo N, Naora H. Molecular Cancer Research.2014;12(4). CrossRef
- Cadherin-6 type 2, K-cadherin (CDH6) is regulated by mutant p53 in the fallopian tube but is not expressed in the ovarian surface Karthikeyan S, Lantvit DD , Chae DH , Burdette JE . Oncotarget.2016;7(43). CrossRef
- Visualizing and interpreting cancer genomics data via the Xena platform Goldman MJ , Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, et al . Nature Biotechnology.2020;38(6). CrossRef
- Gene Expression Analysis in Ovarian Cancer – Faults and Hints from DNA Microarray Study Lisowska KM , Olbryt M, Dudaladava V, Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarząb M, et al . Frontiers in Oncology.2014;4. CrossRef
- GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis Tang Z, Kang B, Li C, Chen T, Zhang Z. Nucleic Acids Research.2019;47(W1). CrossRef
- Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients Győrffy B, Lánczky A, Szállási Z. Endocrine-Related Cancer.2012;19(2). CrossRef
- The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data Cerami E, Gao J, Dogrusoz U, Gross BE , Sumer SO , Aksoy BA , Jacobsen A, et al . Cancer Discovery.2012;2(5). CrossRef
- Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal Gao J, Aksoy BA , Dogrusoz U, Dresdner G, Gross B, Sumer SO , Sun Y, et al . Science Signaling.2013;6(269). CrossRef
- The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL , et al . Nucleic Acids Research.2023;51(D1). CrossRef
- Tissue-based map of the human proteome Uhlén M, Fagerberg L, Hallström BM , Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, et al . Science.2015;347(6220). CrossRef
- GeneMANIA update 2018 Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD , Morris Q. Nucleic Acids Research.2018;46(W1). CrossRef
- Gene Set Knowledge Discovery with Enrichr Xie Z, Bailey A, Kuleshov MV , Clarke DJB , Evangelista JE , Jenkins SL , Lachmann A, et al . Current Protocols.2021;1(3). CrossRef
- GSCALite: a web server for gene set cancer analysis Liu C, Hu F, Xia M, Han L, Zhang Q, Guo A. Bioinformatics.2018;34(21). CrossRef
- E-cadherin expression in human epithelial ovarian cancer and normal ovary Sundfeldt K., Piontkewitz Y., Ivarsson K., Nilsson O., Hellberg P., Brännström M., Janson P. O., et al . International Journal of Cancer.1997;74(3). CrossRef
- E-cadherin expression in ovarian cancer in the laying hen, Gallus domesticus, compared to human ovarian cancer Ansenberger K, Zhuge Y, Lagman JAJ , Richards C, Barua A, Bahr JM , Hales DB . Gynecologic Oncology.2009;113(3). CrossRef
- A comprehensive analysis of different types of databases reveals that CDH1 mRNA and E-cadherin protein are not downregulated in most carcinoma tissues and carcinoma cell lines Sicairos B, Alam S, Du Y. BMC Cancer.2023;23(1). CrossRef
- Epithelial-mesenchymal transitions in development and disease Thiery JP , Acloque H, Huang RYJ , Nieto MA . Cell.2009;139(5). CrossRef
- N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer Mrozik KM , Blaschuk OW , Cheong CM , Zannettino ACW , Vandyke K. BMC Cancer.2018;18(1). CrossRef
- Combined overexpression of cadherin 6, cadherin 11 and cluster of differentiation 44 is associated with lymph node metastasis and poor prognosis in oral squamous cell carcinoma Ma C, Zhao J, Lin R, Zhou L, Chen Y, Yu L, Shi T, et al . Oncology Letters.2018. CrossRef
- N-cadherin haploinsufficiency increases survival in a mouse model of pancreatic cancer Su Y, Li J, Witkiewicz A K, Brennan D, Neill T, Talarico J, Radice G L. Oncogene.2012;31(41). CrossRef
- CDH2 expression is of prognostic significance in glioma and predicts the efficacy of temozolomide therapy in patients with glioblasotma Chen Q, Cai J, Jiang C. Oncology Letters.2018. CrossRef
- Systemic analysis of the expression levels and prognosis of breast cancer-related cadherins Xu M, Liu C, Pu L, Lai J, J, Li J, Ning Q, et al . Experimental Biology and Medicine.2021;246(15):1706-20. CrossRef
- Cadherin switches during epithelial-mesenchymal transition: CDH4/RCAD downregulation reduces bladder cancer progression Martins-Lima C, Miranda-Gonçalves V, Lobo J, Constâncio V, Leite-Silva P, Guimarães-Teixeira C, Monteiro-Reis S, et al . Cellular Oncology.2022;45(1). CrossRef
- N-cadherin inhibitor creates a microenvironment that protect TILs from immune checkpoints and Treg cells Sun Y, Jing J, Xu H, Xu L, Hu H, Tang C, Liu S, et al . Journal for ImmunoTherapy of Cancer.2021;9(3). CrossRef
- An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer Gouin KH , Ing N, Plummer JT , Rosser CJ , Ben Cheikh B, Oh C, Chen SS , et al . Nature Communications.2021;12(1). CrossRef
- E-cadherin germline mutations in familial gastric cancer Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, et al . Nature.1998;392(6674). CrossRef
- Mutation Impact on mRNA Versus Protein Expression across Human Cancers Liu Y, Elmas A, Huang K. 2023. CrossRef
- The miR-200 Family Inhibits Epithelial-Mesenchymal Transition and Cancer Cell Migration by Direct Targeting of E-cadherin Transcriptional Repressors ZEB1 and ZEB2 Korpal M, Lee ES , Hu G, Kang Y. Journal of Biological Chemistry.2008;283(22). CrossRef
- E-Cadherin Mediates MMP Down-Regulation in Highly Invasive Bronchial Tumor Cells Nawrocki-Raby B, Gilles C, Polette M, Martinella-Catusse C, Bonnet N, Puchelle E, Foidart J, Van Roy F, Birembaut P. The American Journal of Pathology.2003;163(2). CrossRef
- Cadherin and catenin alterations in human cancer Hajra KM , Fearon ER . Genes, Chromosomes and Cancer.2002;34(3). CrossRef
- Cadherins: cellular adhesive molecules serving as signalling mediators Yulis M, Kusters DHM , Nusrat A. The Journal of Physiology.2018;596(17). CrossRef
- Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target Yu W, Yang L, Li T, Zhang Y. Frontiers in Oncology.2019;9. CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2025
Author Details