Molecular Analysis of Acquired Tamoxifen Resistance in Breast Cancer Cell Line
Download
Abstract
Background: Tamoxifen is an FDA approved drug for the prevention and the treatment of breast cancer, but its therapeutic benefit is limited by the development of drug resistance. Many Studies suggest that a basic biological difference exists between tumors with acquired tamoxifen resistance and those with intrinsic resistance to the drug. However the reasons why human mammary tumors become resistant to tamoxifen therapy are mainly unknown. Changes in gene expression may occur as cells acquire resistance to tamoxifen. A better understanding of gene expression alterations associated with tamoxifen resistance will facilitate circumventing this problem.
Methods: We undertook a comparative gene expression analysis of tamoxifen-sensitive (MCF7 and T47D) and acquired tamoxifen-resistant human breast cancer cell lines (T47D tamoxifen resistant) in vitro models using Real-time PCR, to analyze differential gene expression. These genes were functionally linked to 4 major groups named as ER signaling/cell cycling, EGFR signaling, cancer stem cell (CSCs) and apoptosis.
Results: Our results have been widely demonstrated that the altered expression of some genes involved in apoptosis, EGFR signaling, CSCs and Cell cycle-independent expression of estrogen receptor such as ESR1, TP53, CDKN1B, Casp3, CD44, CD24, BAX, Bcl2, Her2, and PTEN in T47D tamoxifen resistant cell line can be resulted in drug resistance in long-term treatment.
Conclusions: Our findings correspond with the results of many earlier studies indicated tamoxifen resistance can be a result from combination of molecular mechanisms including Her2 activation, cell cycle progression out of ESR1 control, increased ratio of CSCs and inhibition of mitochondrial apoptosis.
Introduction
Breast cancer is the most commonly diagnosed cancer in women and approximately 70–75% of cases express estrogen receptor alpha (ESR1) [1]. The nonsteroidal anti-estrogen tamoxifen is the drug most often used for the long-term treatment of early breast cancer [2]. Tamoxifen has been shown to be an effective treatment for hormone receptor-positive breast cancer in pre-menopausal and post-menopausal women with all stages of the disease [3, 4]and metabolizes into compounds that bind to the estrogen receptor but do not activate it. Tamoxifen acts by competitive antagonism of estrogen and prevents estrogen from binding to its receptor [5]. This drug provides effective palliation when used to treat patients with advanced disease, and adjuvant tamoxifen therapy produces significant increases in both disease-free and overall survival. Data from the laboratory have shown that the primary action of tamoxifen is tumoristatic rather than tumoricidal, and long-term therapy is therefore recommended [4].
However, about 70–75% of breast tumors express ESR1, and among them 70–80% respond positively to tamoxifen treatment [6], a serious limitation of this endocrine therapy is the inevitable appearance of resistance, which either occurs de novo or acquired resistance. Almost 50% of patients with advanced disease do not respond to first line treatment with tamoxifen [de novo resistance]. The main feature of de novo resistance is the lack of estrogen receptor alpha expression. Moreover, a significant percentage of ESR1 positive breast cancer patients experienced resistance, despite an initial positive drug response [acquired resistance] [7, 8].
To date, many findings suggest that tamoxifen resistance is mediated by several mechanisms including altered expression of ESR1 and/or ESR2, pharmacologic tolerance, emerging of cancer stem cells, EGFR signaling activation and inhibited apoptosis [7, 9, 10].
Since the effects of tamoxifen are primarily mediated through the ESR1, and the degree of ESR1 expression is a strong predictor of responses to tamoxifen, loss of ESR1 expression could confer resistance to tamoxifen therapy [4, 11, 12, 13, 14, 15]. Moreover; it is believed that cancer stem cells may be responsible for resistance against therapeutic approaches and for recurrent tumors. It has been seen that in breast cancer, a population of CD44+/CD24− cells is considered to be highly enriched in cancer-initiating cells which are more tumorigenic than other cell populations [16 ]. In addition, tamoxifen normally acts through the estrogen receptor and down regulates the expression of Her2, but it has been seen that expression of Her2 is up regulated in the presence of tamoxifen, leading to tamoxifen resistant breast cancer [17].
Tamoxifen has been shown to induce apoptosis in breast cancer cells immediately in the absence of Her2 overexpression [18]. One way in which cells potentially acquire resistance to tamoxifen is avoiding drug- induced apoptosis. Resistance to tamoxifen results from a complex series of changes that prevent apoptosis and thus enhance cell proliferation and survival [19].
Whereas many studies focus on anti-estrogen resistance mechanisms, the molecular mechanisms responsible for the development of tamoxifen resistance are unclear [20].
The acquired resistance may be due to changes in specific gene expression involved in different signaling pathways, which eventually could be used as predictive biomarkers of resistance. Furthermore, these markers may be used to select patients that might benefit from additional targeted therapy aside from tamoxifen treatment [21, 22].
In this study, using gene expression profiling of breast cancer cell lines by Real-time PCR, we aim to assess a set of new markers that is predictive for the type of response to endocrine therapy with the tamoxifen. Therefore, we aimed to investigate gene expression of 26 various genes involve in EGFR signaling, ESR signaling/cell cycling, cancer stem cell and apoptosis in MCF7 and T47D cell lines and tamoxien-resistant T47D cell line. These genes are listed in Tabl 1, either a known function or an inferred function was identified from OMIM and GO (Gene Ontology) nomenclature. Gene expression profiling can help to identify predictive markers as well as therapeutic targets in tamoxifen resistant patients.
| Gene symbol | Fold change in T47D TR* versus MCF7 (p value) | Fold change in T47D TR* versus T47D (p value) | Fold change in MCF7 versus T47D (p value) | GO (Gene Ontology) |
| BAX | -5.26 (0.01) | -4.67 (0.005) | -1.13 (0.01) | Induction of apoptosis by extracellular signals |
| BCL2 | 5.56 (0.022) | 2.03 (0.042) | -2.74 (0.003) | Anti-apoptosis; Negative regulation of cell proliferation |
| BCLxL | 1.45 (0.003) | -1.69 (0.013) | -1.17 (0.026) | Apoptosis; Anti-apoptosis |
| BIRC5 | 2.38 (0.016) | 2.67 (0.026) | 1.12 (0.02) | Apoptosis; Anti-apoptosis; Caspase inhibitor activity |
| CASP3 | -2.3 (0.03) | -6.67 (0.01) | 2.9 (0.009) | Apoptosis; Induction of apoptosis |
| CCNB1 | 2.56 (0.02) | 2.2 (0.02) | -1.16 (0.01) | cell cycle regulation; G1/S and G1/M transition of mitotic cell cycle |
| CD24 | -10 (0.001) | -4.12 (0.008) | -2.43 (0.008) | Cancer stem cell marker |
| CD44 | 3.7 (0.019) | 7.69 (0.012) | 2.08 (0.014) | Cancer stem cell marker |
| CDKN1B | -11.11 (0.006) | -5.44 (0.009) | -2.04 (0.01) | Negative regulation of cell proliferation |
| EGFR | 1.61 (0.04) | 1.22 (0.01) | -1.32 (0.023) | EGFR signaling; Epidermal growth factor receptor activity |
| EGR1 | -4.35 (0.042) | 2.33 (0.01) | 1.87 (0.012) | EGFR signaling; Regulation of transcription |
| ESR1 | -3.13 (0.008) | -14.29 (0.007) | 4.57 (0.007) | EGFR signaling; Epidermal growth factor receptor activity |
| ESR2 | -1.69 (0.04) | -7.69 (0.004) | -1.88 (0.008) | Estrogen receptor signaling pathway |
| FLIP | -1.4 (0.023) | -3.67 (0.02) | 2.62 (0.012) | Estrogen receptor signaling pathway |
| Her2 | 5.56 (0.01) | 16.67 (0.014) | 3 (0.014) | Apoptosis; Anti-apoptosis |
| IGF1R | -12.5 (0.04) | -2.86 (0.012) | -4.37 (0.028) | cell cycle regulation; Positive regulation of cell proliferation |
| KRAS | 2.22 (0.04) | -3.88 (0.021) | -1.75 (0.01) | EGFR signaling; Regulation of cell cycle |
| MDM2 | -1.54 (0.054) | -4.26 (0.02) | 2.77 (0.015) | Regulation of cell cycle |
| mTOR | -1.72 (0.023) | -1.45 (0.012) | -1.19 (0.008) | EGFR signaling; positive regulation of cell proliferation |
| NFKB1 | 1.32 (0.028) | 2.78 (0.009) | 2.11 (0.020) | Apoptosis; Anti-apoptosis |
| PGR | -6.25 (0.007) | -12 (0.015) | 1.92 (0.010) | Steroid hormone receptor activity |
| PTEN | -2.54 (0.028) | -7.09 (0.008) | 2.79 (0.01) | EGFR signaling; Negative regulation of cell cycle |
| TP53 | -7.69 (0.018) | -6.33 (0.020) | -1.21 (0.025) | Induction of apoptosis by hormones; Negative regulation of cell growth |
| TRADD | -5.88 (0.028) | -4.29 (0.013) | -1.37 (0.015) | Apoptosis; Anti-apoptosis |
| TRAF2 | -5 (0.02) | -7.45 (0.012) | 1.49 (0.005) | Apoptosis; Induction of apoptosis from TNF receptors |
| VEGF | 1.12 (0.012) | -2.94 (0.019) | -2.63 (0.027) | Positive regulation of cell proliferation; Angiogenesis |
a) T47D-TR: T47D tamoxifen resistant
Materials and Methods
This study was conducted in Pasteur institute of Iran, Tehran, from April 2014 to March 2015.
Cell Culture
The MCF7 and T47D breast cancer cell lines were obtained from the national Cell Bank of IRAN- Pasteur Institute of IRAN. The cells were grown in RPMI-1640 medium (InoClon) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, glutamine and incubation in a tissue culture incubator at 37°C, 5% CO2.
The estrogen sensitive breast cancer cell line T47D was cultured in the presence of tamoxifen (0.1 µM, >5 months). Initially, cell growth rates were very much reduced in the presence of tamoxifen, but eventually cells adapted to the new environment leading to two new sub-lines: T47D tamoxifen resistant and control T47D cells. The T47D tamoxifen resistant derivatives were selected when the initially sensitive cells resumed comparable growth to the parental cells.
The cells were harvest by centrifugation at 1200g for 5 min and the supernatant were decanted and discard.
RNA extraction and cDNA synthesis
For successful and reliable gene expression study, real-time PCR needs high quality, DNA-free, and undegraded RNA, therefore RNA was extracted by RNeasy Mini kit (QIAGEN), residual DNA was removed by on column DNase digestion using the RNase-Free DNase Set (QIAGEN) and the DNase was efficiently removed in subsequent wash steps. After extraction, the quality and yield of extracted RNA was analyzed by UV spectrophotometry using Nanodrop and gel electrophoresis. 1000 ng of total RNA was reverse transcribed using PrimeScript™ II 1st strand cDNA Synthesis Kit (Takara) in reaction volume of 10 µl. As calculated from efficiency plots for each gene, samples of cDNA were diluted 1:5, and 2 µl (20 ng) were used per PCR reaction for all genes.
Quantitative Real-time PCR
Quantitative Real-time PCRs were performed using Maxima SYBR Green qPCR Master Mix (2X) - Thermo Scientific with the Rotor gene 6000 (Corbett) detection system. Primer sequences used for gene expression analysis are listed in Tabl 2. The concentration of primers was optimized to 0.3µM and reactions carried out in triplicate. Designing of primers pairs was optimized so that all primer pairs can work at the same annealing temperature during qPCR. The three Step Real time PCR program used was Hot start at 95°C for 10 min; 40 cycles with denaturation at 95°C for 25 s, annealing at 60°C for 20 s, and extension at 72°C for 15 s. SYBR-green fluorescent signals were used to generate cycle threshold values from which mRNA ratios were calculated when normalized against the ACTB housekeeping gene and represent the log2 mean fold change in expression. The levels of expression of 26 candidate genes including PTEN, CDKN1B, Her2, EGFR, ESR1, PGR, VEGF, NFkB, FLIP, Bcl2, Casp3, TP53, Mdm2, BIRC5, CD44, CD24, KRAS, BclXL, mTOR, BAX, CCNB1, TRADD, TRAF2, IGF1R, EGR1 and ESR2 were investigated. Data were analyzed using the ΔΔCt method, ΔCt values for each sample were calculated by subtracting Ct values from genes of interest from corresponding Ct values of the housekeeping gene ACTB, which exhibited consistent expression across all samples.
| Gene | Description | Forward Primer | Reverse Primer |
| ACTB | Beta-actin, Housekepping gene | GCAAGCAGGAGTATGACGAG | AAGGGTGTAACGCAACTAAGTC |
| BAX | BCL2-associated X protein | CTGTCAGCCACTCTTCTTTCAG | ACAGTAGGTGAGGGTTCTCCAT |
| Bcl2 | B-cell CLL/lymphoma 2 | GATGTGATGCCTCTGCGAAG | CATGCTGATGTCTCTGGAATCT |
| BclXL | BCL2-like 1 | ACCATACTGAGGGACCAACTG | CACAGGCTGCTCTTGTAGGAA |
| BIRC5 | Baculoviral IAP repeat-containing 5 (surviving) | GAGGTCATCTCGGCTGTTCCT | CTGTTGGCAGGCTAGGGAC |
| Casp3 | Caspase 3, apoptosis-related cysteine peptidase | CTCGGTCTGGTACAGATGTCG | CACGCATCAATTCCACAATTTCT |
| CCNB1 | G2/Mitotic-Specific Cyclin B1 | GAGTTACTGAAGGTGATGGAGGTA | ACTAGGGATTCGGTGGTAGACT |
| CD24 | CD24 antigen (small cell lung carcinoma cluster 4 antigen) | ATGGTCACACACTGATGCTTAGA | TGTTCATTCACACACACAGTAGC |
| CD44 | CD44 antigen (Indian blood group) | CCCAGACAGGCTCACTCAA | CAGGTAGAGGCTGTTGTAACCA |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | GGAGCAGACGCCCAAGAAG | GCTTATACAGGATGTCCATTCCA |
| EGFR | Epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | CCCTCCTTACGCTTTGTCAC | GCTACTGTCATTCGCACCTG |
| EGR1 | Early growth response 1 | TGAAACAGCAGTCCCAGTATTC | TGTGAGAGTACGGTCAAGCA |
| Her2 | V-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2), neuro/glioblastoma derived oncogene homolog (avian) | CCAGCAGGGCTTCTTCTGTC | CCAGGTCACCATCAAATACATC |
| ESR1 | Estrogen receptor 1 | CCATCGTCAGTGTGTGTGTTTA | CCAATGACCTCTCTGTGAATG |
| ESR2 | Estrogen receptor 2 (ER beta) | TCTTGTTCTGGACAGGGATGA | TACATACTGGAATTGAGCAGGA |
| FLIP | CASP8 and FADD-like apoptosis regulator | CAAGAACCAGTGAAGAAATCCAT | CCAGGGAAGTGAAGGTGTC |
| IGF1R | Insulin-like growth factor 1 receptor | CGCATCTTTCTGGTCAACAT | CGTCAACTGACTACCCGTAATC |
| KRAS | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | TGCCTGTTTGGGATAATGATAG | ACACAAGACAGTGGAATTGGA |
| Mdm2 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) | CGCTTTATGGGTGGATGCT | CAAGTGCTGGTGCTTTCAGATA |
| mTOR | Mammalian Target Of Rapamycin | CTCACTGGTCGGGACTTCT | CAATATAGCACTGGCAGAGGTT |
| NFkB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | CAAGGCAGCAAATAGACGAG | GTTGAGAGTTAGCAGTGAGGCA |
| PGR | Progesterone receptor | GGTCCCTAAGTACATTGTCCAGA | TGAGGAGCAAGGTGTATGACT |
| PTEN | Phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | GTACCATCCCAAGTCCTTTGTAG | ACCACAGCCATCGTTATGAAC |
| TP53 | Tumor protein p53 (Li-Fraumeni syndrome) | GCTCTGACTGTACCACCATCCAC | GGCACAAACACGCACCTCAAAG |
| TRADD | Tumor necrosis factor receptor type 1-associated DEATH domain protein | TCTGCGGCTATTGCTGAAC | GGAGAAGGTGAGGCTGATCT |
| TRAF2 | TNF receptor-associated factor 2 | CACCAGCCAGTCCTCAGAT | CATCCACTGTGCTCCTGCTA |
| VEGF | Vascular endothelial growth factor | CCTCATCCTCTTCCTGCTC | ACCACTCACACACACACAAC |
Results
EGFR signaling genes
Many previous studies have noted that acquired resistance to tamoxifen is often accompanied by an increased expression of Her2. Because increased expression and signaling via HER2 in breast cancer patients are associated with poor outcome of endocrine therapy, we used HER2 expression as a marker for tamoxifen resistance. Analysis of HER2 levels revealed significant increase in long-term tamoxifen exposure which is reduced sensitivity of breast cancer cells to the antiestrogen tamoxifen (Fig. 1A).
Figure 1A : mRNA expression of 26 genes are compared in T47D-TR*, T47D and MCF7 breast cancer cell lines. These 26 genes were categorized to four groups: a. EGFR signaling:7 genes belong to EGFR signaling involved in regulation of biological process including proliferation, differentiation and survival
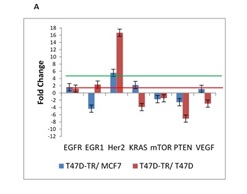
The PTEN gene plays an important role in cell cycle regulation and apoptosis by regulating Protein kinase-B/Akt activity. One of the main objectives of this study was evaluation of PTEN expression, our observation demonstrated reduction in expression of PTEN in T47D tamoxifen resistant cell line in compare to MCF7 and T47D sensitive cell line. In summary our results imply that PTEN can be used as a good prognostic characteristic for the outcome of breast cancer patients treated with tamoxifen.
ESR signaling Genes
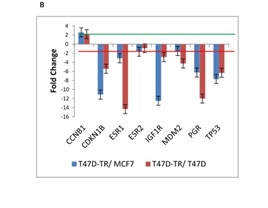
Very prominently, the genes were found to be down-regulated in the T47D tamoxifen-resistant compared with the tamoxifen-sensitive cell lines (T47D and MCF7) were CDKN1B, ESR1, PGR, TP53 and IGF1R, In contrast CCNB1 gene which is involved in G1/S and G1/M transition of mitotic cell cycle is moderately up-regulated in tamoxifen resistant cells (Fig. 1B).
Figure 1B :mRNA expression of 26 genes are compared in T47D-TR*, T47D and MCF7 breast cancer cell lines. These 26 genes were categorized to four groups: b. ESR signaling/Cell cycle .8 genes categorize in ESR signaling involved in cell cycle.
Cancer stem cell
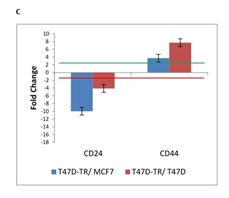
Stem cell theory proposes that cancers arise from malignant transformation of normal stem/progenitor cells. The inherent properties of stem/progenitor cells may impart their transformed counterparts with the ability to evade traditional antitumor therapies and to establish resistance. A subset of human breast cancer cells by a specific cell surface marker profile: the CD44+/CD24- display CSC properties and have an ability to form tumors and also demonstrated a capacity for self-renewal and generation of heterogeneous progeny. Our result indicated up-regulation of CD44 and down-regulation of CD24 in T47D tamoxifen resistant in comparison to T47D sensitive and MCF7 which is implied the activation of cancer stem cell markers (Fig. 1C).
Figure 1C :mRNA expression of 26 genes are compared in T47D-TR*, T47D and MCF7 breast cancer cell lines. These 26 genes were categorized to four groups:c. Cancer stem cell. two genes named as CD24 and CD44 belong to cancer stem cell marker demonstrated a capacity for self-renewal and tumorigenesis
Apoptosis
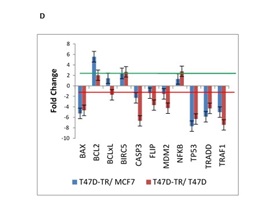
Programmed Cell Death or apoptosis is a highly regulated but not yet fully understood process by which selective gene expression leads to cell demise. Tamoxifen induces cell death in breast cancer cells via apoptosis. It is effective in breast cancer prevention and treatment by inhibiting the proliferative effects of estrogen that are mediated through the estrogen receptor (ER). Its principal mechanism of action is stopping the proliferation of the cells that is initiated by estrogen stimulation (cytostatic); this action is mediated by the drug competing with estrogen and subsequently binding to the estrogen receptor in mammary epithelia to form a nuclear complex that decreases DNA binding and estrogenic effects. To understand the changes in tamoxifen induced apoptosis in breast cancer cells, we investigated the expression profiles of 11 apoptosis related genes. Our data showed long-term tamoxifen treatment of cell lines decreased levels of several pro-apoptotic genes including TRAF2, CASP3, BAX and TP53, whereas the anti-apoptotic members, which include Bcl2 and BIRC5 were up-regulated in T47D tamoxifen resistance compared to T47D and MCF7.
The genes BAX, FLIP, TP53 and TRAF2 was found to be down-regulated in T47D tamoxifen resistance treated permanently with tamoxifen compared with the sensitive T47D cells. In general, our expression patterns indicate that tamoxifen resistance is mainly associated with inhibition of apoptosis through mitochondrial pathway (Fig. 1D).
Figure 1D : mRNA expression of 26 genes are compared in T47D-TR*, T47D and MCF7 breast cancer cell lines. These 26 genes were categorized to four groups: d. Apoptosis. 11 genes belong to apoptosis regulation involved in programmed cell death.
Discussion
Breast cancer is the most frequently diagnosed female carcinoma and the second leading cause of cancer death for women of all ages.
The antiestrogen tamoxifen is the usual endocrine therapy for estrogen-related breast cancer patient. Use of tamoxifen for five years reduces the annual relapse rate by 39% and the annual mortality rate by 31% [23].
Despite the benefits of tamoxifen in treatment of ESR1 positive breast cancer, almost all patients with metastatic disease and about 40% of patients receiving adjuvant tamoxifen eventually relapse and die from breast cancer. The biological mechanisms underlying de novo and acquired resistance to tamoxifen are still a challenging clinical problem, and the mechanisms underlying tamoxifen resistance, which probably develops through multiple pathways, are still unclear. Understanding of these mechanisms may suggest novel strategies to overcome tamoxifen resistance and make further improvements in breast cancer survival [7].
In present study, we examined the mechanism of tamoxifen resistance through expression study of 26 genes in four categories including, ESR signaling/cell cycle, EGFR signaling, cancer stem cell and apoptosis.
Previous studies have indicated that acquired resistance to tamoxifen is occurred for some events such as Loss of ESR1 expression/function, altered expression of ESR2, pharmacologic tolerance, alterations in ESR1 co-activator proteins, alterations in ESR1 co-repressor proteins, altered expression of EGFR or Her2 and altered expression/function of IGF1R [7].
Pharmacologic tolerance is a common mechanism of drug resistance such as the decrease of intracellular concentrations of drug as a result of decreased influx or increased efflux; however the drug efflux is often mediated by the membrane pump P-glycoprotein, the mechanism responsible for altered tamoxifen accumulation is not understood [11]. Sensitivity to tamoxifen could also be affected by a reduction in the intracellular availability of tamoxifen due to the presence of intracellular antiestrogen binding sites [24]. The extent to which such a mechanism contributes to clinical resistance is not known. The fact that tamoxifen and its metabolites saturate the ESR1 by more than 99.9% in post-menopausal women suggests that only exceptionally large changes in the disposition of tamoxifen could lead to resistance [25]. Increased metabolism of tamoxifen to agonistic metabolites is a further potential resistance mechanism [26]. Tamoxifen is metabolized to N-desmethyltamoxifen and 4-hydroxytamoxifen. N-desmethyltamoxifen is the major metabolite detected in the serum; despite 4-hydroxytamoxifen has a greater affinity for the ESR1 than tamoxifen; the levels in serum are very low. The pharmacogenetics studies indicate that both pharmacogenomic effects and pharmacological interactions may alter the metabolism and potentially the efficacy of tamoxifen [27, 28].
Since the effects of tamoxifen are mediated through the ESR1, loss of ESR1 expression could confer resistance to endocrine therapy [4, 11, 12, 13, 14, 15]. Furthermore, ESR1 mutations may lead to a loss of function of ESR1 without alteration of its expression [29, 30, 31].
ESR2 have several variant forms and is expressed in both normal and malignant breast tissue [32, 33, 34]. ESR2 is highly homologous to ESR1 in its DNA and binds oestradiol with similar affinity to ESR1 [35, 36, 37, 38, 39, 40]. Speirs et al had reported that using RT-PCR, median ESR2 mRNA levels were approximately 2-fold higher than ESR1 levels in tamoxifen-resistant tumors compared with tamoxifen- sensitive tumors [41].
Our finding was shown the level of ESR1 gene expression significantly down regulated in tamoxifen-resistant T47D versus both T47D and MCF7 cell line. The comparison of gene expression profiles between the tamoxifen-resistant T47D cell line and the estrogen-stimulated T47D and MCF7 clearly showed while the ESR1, PGR and IGF1R expression as cell cycle stimulators were down regulated, the cell cycle negative regulators including CDKN1B and TP53 have also been reduced. On the other hand, CCNB1 as regulator of G1/S and G1/M transition was up regulated. It seems in tamoxifen-resistant cell lines, cell cycle is continued out of ESR signaling control. Therefore, genes that control the cell cycle have the potential to consequence drug sensitivity and resistance.
Similarly, a recent study indicated over-expression of CCND1, a direct transcriptional target of ESR signaling, conferred resistance to tamoxifen in vitro and was up-regulated in de novo tamoxifen-resistant cells [42, 43].
Clinical data demonstrate that tamoxifen resistant breast cancers often have an increased expression of Her2 and/or EGFR [44, 45, 46]. Also other data suggest a role for ESR1 phosphorylation by EGFR downstream signaling, in anti-estrogen resistance [47, 48, 49, 50, 51]. Since MAPK can be activated downstream from EGFR and/or Her2 and leading to increased phosphorylation of the ESR1 on serine 118 or serine 167, together these observations suggest that the EGFR/ Her2 signaling pathways might play a role in tamoxifen resistance [52, 53, 54]. Therefore, we investigated the effect of EGFR signaling pathway on tamoxifen resistance. In this study as expected, tamoxifen greatly increased the HER2 17 fold up-regulated in T47D TamRes versus T47D. Therefore, it seems increased HER2 expression in ESR1 positive breast cancer cell lines may be an important determinant for prediction of endocrine resistance. Our data are consistent with literature data showing tamoxifen resistance upon increased HER2 expression in breast cancer cells, typically those studies involved human breast cancer cell lines that were long term cultured in the presence of tamoxifen. Furthermore, some studies have reported that HER2 or EGFR expression are associated with the development of tamoxifen resistance and it could be moderated by inhibition of HER2 or EGFR in tamoxifen resistance cells [55, 56].
On the other hand previous studies have shown that PTEN down-regulation in breast cancer is associated with high-grade tumor, distant metastases and poorer disease-free survival. This gene is involved in the regulation of the cell cycle, preventing cells from growing and dividing too rapidly. Decreased PTEN in breast cancer cells has recently been associated with resistance to tamoxifen-induced apoptosis. Previous studies reported an association between reduced (or loss of) PTEN expression and advanced breast cancer stage, suggesting that loss of PTEN expression might have prognostic relevance and their results in relation to outcome of the patients and association of reduced PTEN expression with adverse clinic pathological variables were similar [57]. In this study, we found 9-fold down-regulation of PTEN expression in tamoxifen resistance T47D versus T47D. Our data, in combination with previous works, suggests that the reduced PTEN expression is significantly associated with tamoxifen resistance.
The originating of cancer stem cells from various cancers is comparatively resistant to commonly used anti cancer drugs [58, 59, 60, 61, 62]. There are several suggested mechanisms for the apparent resistance of CSCs to current anticancer therapies. a) Slow growth of CSCs and therefore resistance to anti cancer drugs adapted to fast growing cancer cells, b) Increased activation of the response to DNA damage checkpoint, c) Enhanced expression of efflux transporters, these transporters efflux anti cancer drugs, which is a common cause of drug resistance [63, 64, 65]. In this study, The CD44/CD24 subpopulation ratio and its effect on tamoxifen were analyzed in MCF-7, T47D and T47D tamoxifen resistant cells. Surprisingly, we found that tamoxifen resistance cells expressed a much higher and lower levels of CD44 and CD24, respectively (CD44+/CD24-). As our result, it can be suggested that the CD44/CD24 subpopulation ratio in tamoxifen resistance cells is higher than that in MCF-7 and T47D cells, indicating that acquired tamoxifen resistance might be associated with the ratio of breast cancer stem cells. This may be especially true for tumors that are composed of a heterogeneous population of cancer cells.
Tamoxifen has been shown to induce apoptosis in breast cancer cells and tumor cell evasion of apoptosis contributes to tamoxifen resistance [66, 67].
Several studies have demonstrated that over expression of the Bcl2 confers resistance to apoptosis by apoptotic inducers in several cancers [68, 69, 70]. In breast cancers, high level of Bcl2 expression has been shown to be a favorable prognosis of response to endocrine therapy [4, 11, 71]. This desirable response to endocrine therapy by Bcl2-positive tumors might be the result of down-regulation of Bcl2 by endocrine therapeutic agents such as tamoxifen, with the consequent susceptibility to apoptosis [72]. We find results indicating up-regulation of Bcl2 as anti-apoptotic factor and down-regulation of BAX, TP53 and Casp3, which are pro-apoptotic factors.
The Bcl2:bax ratio is important in determining whether a cell will undergo apoptosis or survive [73]. Our results demonstrated increasing of Bcl2/Bax ratio which is important in cell surviving. TP53 performs a critical role in apoptosis after treatment with anti cancer drugs by down-regulation of Bcl2 [72]. Since TP53 expression is decreased in T47D tamoxifen resistant cells, our results suggest that loss of TP53 and over-expression of Bcl2 compromise tamoxifen therapy. With respect to these observations, it seems that the mitochondrial pathway of apoptosis or intrinsic pathway was inhibited in tamoxifen resistance and plays the critical role in tumor regression.
In short, we found tamoxifen resistance can be a result from combination of molecular mechanisms including Her2 activation, cell cycle progression out of ESR1 control, increased ratio of CSCs and inhibition of mitochondrial apoptosis. Our findings correspond with the \results of many earlier studies, it seems it is time to make a clinical decision to decrease breast cancer relapse due to tamoxifen resistance.
Finally, we suggest for clinical evaluation of molecular markers beside to ESR1 assessment to see whether these markers can be used to predict the likelihood of resistance. With respect to gene expression analysis it can be estimate how likely cancer cells are to respond to tamoxifen, which patients might benefit from another drug along with tamoxifen treatment and who needs adjuvant therapy.
References
[1]. Piva M, Domenici G, Iriondo O, Rabano M, Simoes BM, Comaills V, et al. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol Med. 2014;6(1):66-79.
[2]. Ponzone R, Biglia N, Jacomuzzi ME, Mariani L, Dominguez A, Sismondi P. Antihormones in prevention and treatment of breast cancer. Ann N Y Acad Sci. 2006;1089:143-58.
[3]. Group BIGC, Mouridsen H, Giobbie-Hurder A, Goldhirsch A, Thurlimann B, Paridaens R, et al. Letrozole therapy alone or in sequence with tamoxifen in women with breast cancer. N Engl J Med. 2009;361(8):766-76.
[4]. Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1-22.
[5]. de Cremoux P, Dieras V, Poupon MF, Magdelenat H, Sigal-Zafrani B, Fourquet A, et al. [Tamoxifen and aromatase inhibitors in the treatment of breast cancer in menopausal women: pharmacological and clinical aspects]. Bull Cancer. 2004;91(12):917-27.
[6]. Badia E, Oliva J, Balaguer P, Cavailles V. Tamoxifen resistance and epigenetic modifications in breast cancer cell lines. Curr Med Chem. 2007;14(28):3035-45.
[7]. Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11(4):643-58.
[8]. Garcia-Becerra R, Santos N, Diaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci. 2012;14(1):108-45.
[9]. Ferrer-Soler L, Vazquez-Martin A, Brunet J, Menendez JA, De Llorens R, Colomer R. An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa) -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review). Int J Mol Med. 2007;20(1):3-10.
[10]. Roskoski R, Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34-74.
[11]. Rastelli F, Crispino S. Factors predictive of response to hormone therapy in breast cancer. Tumori. 2008;94(3):370-83.
[12]. von Minckwitz G, Sinn HP, Raab G, Loibl S, Blohmer JU, Eidtmann H, et al. Clinical response after two cycles compared to HER2, Ki-67, p53, and bcl-2 in independently predicting a pathological complete response after preoperative chemotherapy in patients with operable carcinoma of the breast. Breast Cancer Res. 2008;10(2):R30.
[13]. Hamilton N, Marquez-Garban D, Mah V, Fernando G, Elshimali Y, Garban H, et al. Biologic roles of estrogen receptor-beta and insulin-like growth factor-2 in triple-negative breast cancer. Biomed Res Int. 2015;2015:925703.
[14]. Ingle JN, Suman VJ, Mailliard JA, Kugler JW, Krook JE, Michalak JC, et al. Randomized trial of tamoxifen alone or combined with fluoxymesterone as adjuvant therapy in postmenopausal women with resected estrogen receptor positive breast cancer. North Central Cancer Treatment Group Trial 89-30-52. Breast Cancer Res Treat. 2006;98(2):217-22.
[15]. Mariotto AB, Feuer EJ, Harlan LC, Abrams J. Dissemination of adjuvant multiagent chemotherapy and tamoxifen for breast cancer in the United States using estrogen receptor information: 1975-1999. J Natl Cancer Inst Monogr. 2006;(36):7-15.
[16]. Yenigun VB, Ozpolat B, Kose GT. Response of CD44+/CD24-/low breast cancer stem/progenitor cells to tamoxifen and doxorubicininduced autophagy. Int J Mol Med. 2013;31(6):1477-83.
[17]. Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456(7222):663-6.
[18]. Parra-Palau JL, Pedersen K, Peg V, Scaltriti M, Angelini PD, Escorihuela M, et al. A major role of p95/611-CTF, a carboxy-terminal fragment of HER2, in the down-modulation of the estrogen receptor in HER2-positive breast cancers. Cancer Res. 2010;70(21):8537-46.
[19]. Lewis-Wambi JS, Kim HR, Wambi C, Patel R, Pyle JR, Klein-Szanto AJ, et al. Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis. Breast Cancer Res. 2008;10(6):R104.
[20]. Sun X, Mao Y, Wang J, Zu L, Hao M, Cheng G, et al. IL-6 secreted by cancer-associated fibroblasts induces tamoxifen resistance in luminal breast cancer. Oncogene. 2014.
[21]. Beelen K, Zwart W, Linn SC. Can predictive biomarkers in breast cancer guide adjuvant endocrine therapy? Nat Rev Clin Oncol. 2012;9(9):529-41.
[22]. Hortobagyi GN. Toward individualized breast cancer therapy: translating biological concepts to the bedside. Oncologist. 2012;17(4):577-84.
[23]. Early Breast Cancer Trialists' Collaborative G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687-717.
[24]. Chang M. Tamoxifen resistance in breast cancer. Biomol Ther (Seoul). 2012;20(3):256-67.
[25]. Saladores P, Murdter T, Eccles D, Chowbay B, Zgheib NK, Winter S, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2015;15(1):84-94.
[26]. Selever J, Gu G, Lewis MT, Beyer A, Herynk MH, Covington KR, et al. Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clin Cancer Res. 2011;17(20):6510-21.
[27]. Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40.
[28]. Singh MS, Francis PA, Michael M. Tamoxifen, cytochrome P450 genes and breast cancer clinical outcomes. Breast. 2011;20(2):111-8.
[29]. Gu Y, Chen T, Lopez E, Wu W, Wang X, Cao J, et al. The therapeutic target of estrogen receptor-alpha36 in estrogen-dependent tumors. J Transl Med. 2014;12:16.
[30]. Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20(7):1757-67.
[31]. Fan P, Craig Jordan V. Acquired resistance to selective estrogen receptor modulators (SERMs) in clinical practice (tamoxifen & raloxifene) by selection pressure in breast cancer cell populations. Steroids. 2014;90:44-52.
[32]. Al-Bader M, Ford C, Al-Ayadhy B, Francis I. Analysis of estrogen receptor isoforms and variants in breast cancer cell lines. Exp Ther Med. 2011;2(3):537-44.
[33]. Beato M, Vicent GP. A new role for an old player: steroid receptor RNA Activator (SRA) represses hormone inducible genes. Transcription. 2013;4(4):167-71.
[34]. Maehle BO, Collett K, Tretli S, Akslen LA, Grotmol T. Estrogen receptor beta--an independent prognostic marker in estrogen receptor alpha and progesterone receptor-positive breast cancer? APMIS. 2009;117(9):644-50.
[35]. Tonetti DA, Gao W, Escarzaga D, Walters K, Szafran A, Coon JS. PKCalpha and ERbeta Are Associated with Triple-Negative Breast Cancers in African American and Caucasian Patients. Int J Breast Cancer. 2012;2012:740353.
[36]. Bozkurt KK, Kapucuoglu N. Investigation of immunohistochemical ERalpha, ERbeta and ERbetacx expressions in normal and neoplastic breast tissues. Pathol Res Pract. 2012;208(3):133-9.
[37]. Novelli F, Milella M, Melucci E, Di Benedetto A, Sperduti I, Perrone-Donnorso R, et al. A divergent role for estrogen receptor-beta in node-positive and node-negative breast cancer classified according to molecular subtypes: an observational prospective study. Breast Cancer Res. 2008;10(5):R74.
[38]. Hussein MR, Abd-Elwahed SR, Abdulwahed AR. Alterations of estrogen receptors, progesterone receptors and c-erbB2 oncogene protein expression in ductal carcinomas of the breast. Cell Biol Int. 2008;32(6):698-707.
[39]. Reese JM, Suman VJ, Subramaniam M, Wu X, Negron V, Gingery A, et al. ERbeta1: characterization, prognosis, and evaluation of treatment strategies in ERalpha-positive and -negative breast cancer. BMC Cancer. 2014;14:749.
[40]. Guo L, Hou X, Dilimina Y, Wang B. Expression of ERbeta, ERalpha and Her-2 and distribution of molecular subtypes in Uygur and Han patients with breast cancer. Exp Ther Med. 2014;7(5):1077-82.
[41]. Gryder BE, Rood MK, Johnson KA, Patil V, Raftery ED, Yao LP, et al. Histone deacetylase inhibitors equipped with estrogen receptor modulation activity. J Med Chem. 2013;56(14):5782-96.
[42]. Ward A, Shukla K, Balwierz A, Soons Z, Konig R, Sahin O, et al. MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J Pathol. 2014;233(4):368-79.
[43]. Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68(3):826-33.
[44]. Gee JM, Robertson JF, Gutteridge E, Ellis IO, Pinder SE, Rubini M, et al. Epidermal growth factor receptor/HER2/insulin-like growth factor receptor signalling and oestrogen receptor activity in clinical breast cancer. Endocr Relat Cancer. 2005;12 Suppl 1:S99-S111.
[45]. Moerkens M, Zhang Y, Wester L, van de Water B, Meerman JH. Epidermal growth factor receptor signalling in human breast cancer cells operates parallel to estrogen receptor alpha signalling and results in tamoxifen insensitive proliferation. BMC Cancer. 2014;14:283.
[46]. Giuliano M, Trivedi MV, Schiff R. Bidirectional Crosstalk between the Estrogen Receptor and Human Epidermal Growth Factor Receptor 2 Signaling Pathways in Breast Cancer: Molecular Basis and Clinical Implications. Breast Care (Basel). 2013;8(4):256-62.
[47]. Ghayad SE, Vendrell JA, Ben Larbi S, Dumontet C, Bieche I, Cohen PA. Endocrine resistance associated with activated ErbB system in breast cancer cells is reversed by inhibiting MAPK or PI3K/Akt signaling pathways. Int J Cancer. 2010;126(2):545-62.
[48]. Cheng J, Zhang C, Shapiro DJ. A functional serine 118 phosphorylation site in estrogen receptor-alpha is required for down-regulation of gene expression by 17beta-estradiol and 4-hydroxytamoxifen. Endocrinology. 2007;148(10):4634-41.
[49]. Likhite VS, Stossi F, Kim K, Katzenellenbogen BS, Katzenellenbogen JA. Kinase-specific phosphorylation of the estrogen receptor changes receptor interactions with ligand, deoxyribonucleic acid, and coregulators associated with alterations in estrogen and tamoxifen activity. Mol Endocrinol. 2006;20(12):3120-32.
[50]. Zwart W, Griekspoor A, Rondaij M, Verwoerd D, Neefjes J, Michalides R. Classification of anti-estrogens according to intramolecular FRET effects on phospho-mutants of estrogen receptor alpha. Mol Cancer Ther. 2007;6(5):1526-33.
[51]. Britton DJ, Hutcheson IR, Knowlden JM, Barrow D, Giles M, McClelland RA, et al. Bidirectional cross talk between ERalpha and EGFR signalling pathways regulates tamoxifen-resistant growth. Breast Cancer Res Treat. 2006;96(2):131-46.
[52]. Derin D, Eralp Y, Ozluk Y, Yavuz E, Guney N, Saip P, et al. Lower level of MAPK expression is associated with anthracycline resistance and decreased survival in patients with hormone receptor negative breast cancer. Cancer Invest. 2008;26(7):671-9.
[53]. Sarwar N, Kim JS, Jiang J, Peston D, Sinnett HD, Madden P, et al. Phosphorylation of ERalpha at serine 118 in primary breast cancer and in tamoxifen-resistant tumours is indicative of a complex role for ERalpha phosphorylation in breast cancer progression. Endocr Relat Cancer. 2006;13(3):851-61.
[54]. Yamashita H, Nishio M, Kobayashi S, Ando Y, Sugiura H, Zhang Z, et al. Phosphorylation of estrogen receptor alpha serine 167 is predictive of response to endocrine therapy and increases postrelapse survival in metastatic breast cancer. Breast Cancer Res. 2005;7(5):753-64.
[55]. Kim YS, Won YS, Park KS, Song BJ, Kim JS, Oh SJ, et al. Prognostic significance of HER2 gene amplification according to stage of breast cancer. J Korean Med Sci. 2008;23(3):414-20.
[56]. Candas D, Lu CL, Fan M, Chuang FY, Sweeney C, Borowsky AD, et al. Mitochondrial MKP1 is a target for therapy-resistant HER2-positive breast cancer cells. Cancer Res. 2014;74(24):7498-509.
[57]. Shoman N, Klassen S, McFadden A, Bickis MG, Torlakovic E, Chibbar R. Reduced PTEN expression predicts relapse in patients with breast carcinoma treated by tamoxifen. Mod Pathol. 2005;18(2):250-9.
[58]. Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1(4):389-402.
[59]. Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67.
[60]. Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27(12):1749-58.
[61]. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313-23.
[62]. Schulenburg A, Blatt K, Cerny-Reiterer S, Sadovnik I, Herrmann H, Marian B, et al. Cancer stem cells in basic science and in translational oncology: can we translate into clinical application? J Hematol Oncol. 2015;8(1):16.
[63]. Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267-84.
[64]. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756-60.
[65]. Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101(3):781-6.
[66]. Girard BJ, Regan Anderson TM, Welch SL, Nicely J, Seewaldt VL, Ostrander JH. Cytoplasmic PELP1 and ERRgamma protect human mammary epithelial cells from Tam-induced cell death. PLoS One. 2015;10(3):e0121206.
[67]. Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE. The HER4/4ICD estrogen receptor coactivator and BH3-only protein is an effector of tamoxifen-induced apoptosis. Cancer Res. 2008;68(15):6387-95.
[68]. Woo KJ, Yoo YH, Park JW, Kwon TK. Bcl-2 attenuates anticancer agents-induced apoptosis by sustained activation of Akt/protein kinase B in U937 cells. Apoptosis. 2005;10(6):1333-43.
[69]. Raisova M, Goltz G, Bektas M, Bielawska A, Riebeling C, Hossini AM, et al. Bcl-2 overexpression prevents apoptosis induced by ceramidase inhibitors in malignant melanoma and HaCaT keratinocytes. FEBS Lett. 2002;516(1-3):47-52.
[70]. Wang Y, Wang X, Zhao H, Liang B, Du Q. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J Chemother. 2012;24(6):348-57.
[71]. Lee KH, Im SA, Oh DY, Lee SH, Chie EK, Han W, et al. Prognostic significance of bcl-2 expression in stage III breast cancer patients who had received doxorubicin and cyclophosphamide followed by paclitaxel as adjuvant chemotherapy. BMC Cancer. 2007;7:63.
[72]. Bogush T, Dudko E, Bogush E, Polotsky B, Tjulandin S, Davydov M. Tamoxifen non-estrogen receptor mediated molecular targets. Oncol Rev. 2012;6(2):e15.
[73]. Mohamad N, Gutierrez A, Nunez M, Cocca C, Martin G, Cricco G, et al. Mitochondrial apoptotic pathways. Biocell. 2005;29(2):149-61.
License
Copyright
© ,
Author Details