HLA-G: Facts and Fictions
Download
Abstract
Human leukocyte antigen (HLA)-G is a nonclassical MHC class I molecule with modulatory effects on NK and T cells. Unlike classical HLA class I molecules, HLA-G has seven isoforms, three of which are soluble. Soluble HLA-G molecules are reportedly able to transduce negative signals to immune cells after interacting with their corresponding receptors. The expression of these molecules plays significant roles in maternal tolerance against semi-allogenic fetuses. Overexpression of HLA-G in tumors and increased serum levels of soluble HLA-G have been reported in different malignancies, and these changes may be involved in tumoral immune evasion and cancer progression. To improve immune responses against tumor cells, the downmodulation of HLA-G by siRNA or blocking monoclonal antibodies can be helpful in cancer immunotherapy. Additionally, HLA-G can be considered a potential biomarker for the diagnosis and/or prognosis of certain cancers. Although polymorphism of the HLA-G gene-coding region is more limited than in classical HLA class I, some genetic variations in regulatory regions of the gene control the expression level of this molecule. Furthermore, epigenetic factors such as infections may affect the expression of HLA-G in infection-related cancers.
HLA-G: from gene to protein
HLA-G is a nonclassical class I MHC molecule encoded by a gene on chromosome 6, downstream from the classical HLA class I genes. It contains eight exons which encode signal peptide, α1, α2 and α3 domains, and the transmembrane and cytoplasmic region. Due to the presence of a stop codon in exon 6, the last two exons are not translated. To date, the 58 alleles reported for this gene are believed to encode only 18 different proteins (http://hla.alleles.org/nomenclature/stats.html), because most exonic variations are synonymous and the remaining variation sites are intronic or located in regulatory regions [1].
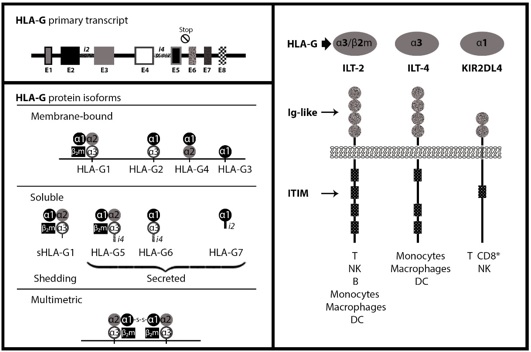
HLA-G generates seven different isoforms through alternative splicing. Four of them are membrane-bound (G1-G4) and three isoforms are soluble forms (G5-G7). HLA-G1 contains a complete α chain with three domains and is expressed in combination with β2-microglobulin. HLA-G2 contains the α1 and α3 domains, HLA-G3 contains the α1 and α2 domains, and HLA-G4 consists of a single α1 domain. HLA-G5, -G6 and -G7 are soluble analogues of HLA-G1, -G2 and -G4, respectively (Fig.1) [2]. Due to the existence of intron-4 or -2 in the mRNA of soluble isoforms which contain the stop codon, the transmembrane and cytoplasmic domains of HLA-G5, -G6 and -G7 are not translated [1]. Soluble HLA-G (sHLA-G) can also be generated through proteolytic cleavage of membrane-bound isoforms. HLA-G can also form dimers via disulfide bonds through Cys-42 in the α1 domain, which enhance the avidity of the molecule for its corresponding receptors [2].
Figure 1 :HLA-G gene and transcription products
HLA-G receptors
HLA-G interacts with immune cells through multiple receptors such as immunoglobulin-like transcript (ILT)2, ILT4, and killer-cell immunoglobulin-like receptor (KIR(2DL4. ILT2 is expressed by natural killer (NK) cells, T cells, dendritic cells (DCs) and decidual macrophages. Expression of ILT4 is limited to myeloid cells and DCs, whereas KIR2DL4 is expressed in all NK cells [3]. Although these receptors can bind both classical and nonclassical HLA class I molecules, they have higher affinity for HLA-G than classical HLA class I molecules. Moreover, in contrast to classical HLA class I, the interaction of HLA-G molecules on tumor cells with ILT2 on NK cells does not require tumor cell lipid raft integrity [4].
ILT2 and ILT4 possess four and three immune receptor tyrosine-based inhibitory motifs (ITIMs), respectively, in their long cytoplasmic tails. These receptors contain four extracellular domains, D1–D4, and mediate the interaction of these receptors with HLA class I molecules. However, KIR2DL4 has a long cytoplasmic tail, like inhibitory KIRs, and also has a charged amino acid (arginine) in the transmembrane domain which can act to activate KIRs. It has been shown that because of conformational prevention, KIR2DL4 cannot bind HLA-G dimers [3].
Another recognized HLA-G receptor is CD160, expressed by endothelial cells. Engagement of the CD160 receptor by HLA-G triggers the apoptosis of endothelial cells and inhibits their proliferation and the angiogenic process [5].
HLA-G immune function
HLA-G exerts immunomodulatory functions via direct and indirect mechanisms. HLA-G interacts with inhibitory receptors on NK cells and cytotoxic T lymphocytes (CTLs), leading to their functional inhibition [6]. It has been shown that sHLA-G is able to induce apoptosis in CTLs [7]. HLA-G induces upregulation of inhibitory receptors, including itself [8], and also has indirect inhibitory functions through the generation of regulatory cells. HLA-G-induced regulatory cells require HLA-G for their generation but not for their function [9]. For example, HLA-G1-transfected antigen-presenting cells (APCs) lead to CD4+ and CD8+ T cell unresponsiveness to alloantigens, and allow them to acquire a regulatory phenotype [10]. Another indirect immunomodulatory effect of HLA‑G arises from HLA-G and HLA-E cooperation. HLA‑E directly binds to HLA‑G-derived signal peptide, and then interacts with CD94/NKG2A, the inhibitory receptor on NK cells [11].
The HLA-G story
HLA-G was discovered by Geraghty et al. in 1987 [12], and its modulatory functions in the trophoblast during pregnancy were initially described in 1990 [13]. Semi-allogeneic fetuses evade rejection by maternal leukocytes via different mechanisms such as expressing Fas ligand (FasL), tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), indoleamine 2,3-dioxygenase (IDO) and programmed death-ligand 1 (PD-L1). In addition to these immunomodulatory molecules, the extravillous trophoblast (EVT) which invades the maternal uterine mucosa lacks HLA-A and HLA-B molecules, but expresses HLA-C, -E and -G [14, 15]. HLA-G was first introduced as a key molecule for the maintenance of maternal tolerance by NK cell inhibition [16]. The trophoblast is recognizable by NK cells thanks to the absence of HLA class I molecules, but HLA-G expression on the trophoblast helps this tissue to avoid NK cell-mediated lysis [15]. The immunosuppressive effects of HLA-G were supported by the detection of decidual DC-10, a major APC subset in decidua, which expresses high levels of HLA-G which in turn induce ILT4 on infiltrating immune cells and maintain tolerance during pregnancy [17].
In addition to protecting the fetus from maternal immune cell attack, HLA-G expression on EVT plays an important role in uterine vascular remodeling. Unlike peripheral NK cells, decidual NK cells (dNK) are less cytotoxic and mostly secrete cytokines and growth factors such as IFN-γ, TGF-β, VEGF, placental growth factor (PLGF) and angiopoietin 1/2 [18]. Moreover, β2m-bound HLA-G isoforms are not expressed on distal EVT, and these invasive cells express only heavy chain-containing HLA-G isoforms which may have limited inhibitory effect because they are not recognized by ILT2 [19, 20]. Decidual NK cells, which express more KIR2DL4 than ILT2, secret IL-6, IL-8 and TNF-α after interaction with HLA-G, and this subsequently leads to NK cell-mediated EVT invasion into the decidua [21]. Therefore, HLA-G can play potentially important roles in maternal tolerance of the fetus through different mechanisms.
Aberrant HLA-G expression has been observed under nonphysiological conditions such as viral infection, cancer, transplantation, and in inflammatory and autoimmune diseases. HLA-G may have two distinct effects in pathological conditions: it can be useful in inflammatory and autoimmune diseases, or dangerous in tumors and infections [3]. In many studies, increased HLA-G expression was shown to be associated with graft acceptance and the prevention of chronic rejection [22].
HLA-G and cancer
Malignant cells typically develop mechanisms to evade immune surveillance [23]. HLA class I downregulation and HLA-G overexpression play critical roles in the modulation of immune responses against tumors. Because tumor cells express mutated or normal but downregulated antigens, they can be recognized as missing-self, non-self and stress-induced self by the immune system. This is the first step in the immune editing mechanism called the elimination phase. In this primary phase, most tumor cells are recognized by the immune system and eliminated by NK cells and CTLs. When a tumor is not eradicated completely, resistant variants may be selected: this mechanism selects cells that are less immunogenic, and gives rise to the equilibrium phase. When immune-resistant variants are maintained and expanded, the evasion phase begins [24, 25]. HLA-G molecules play crucial roles in all phases of immune editing by inhibiting the cytolytic action of NK cells and CTLs [7, 26]. HLA-G-positive tumors might progress from the elimination phase to the equilibrium phase through NK cell inhibition [26, 27]. These HLA-G-positive cells are less immunogenic and more resistant to immune system recognition and cytolysis. Therefore they are positively selected and lead to the evasion phase, characterized by rapid tumor growth [24].
HLA-G was shown to be expressed by melanoma cells in 1998 [28]. In the following years, HLA-G expression was frequently detected in different solid tumors [29, 30, 31]. What emerged from these studies was that HLA-G could potentially be expressed by all tumor types. Although HLA-G is mostly expressed by tumor cells rather than the surrounding healthy tissues [32], expression levels were reported to be highly variable (from 0% to 100%) in different tumors. Moreover, plasma levels of sHLA-G are significantly higher in patients with different malignancies than in healthy controls [33, 34, 35].
Many recent studies have assessed the clinical relevance of HLA-G expression in different cancers, as summarized in Tabl 1. In general, high HLA-G expression correlates with advanced disease stages, poor histological grade, metastases, shorter survival times and tumor recurrence [29, 30, 40, 43]. It was also previously demonstrated that HLA-G overexpression in solid tumors correlates positively with the number of infiltrating Treg cells, and negatively with CD8-positive T cell and NK cell activities [44, 45]. No association has been reported between increased HLA-G expression and clinicopathological aspects in malignancies such as bladder cancer [46] and acute myeloid leukemia [47]. However, we recently found a correlation between HLA-G overexpression in tumor tissues and advanced tumor stages and grades in pancreatic adenocarcinoma, although we found no correlation between HLA-G expression and clinicopathological parameters in hepatocellular carcinoma (HCC) [48].
| Cancer type | Methods | Observations | Main conclusion | Ref |
| Gastric cancer | FCM ELISA | The number of HLA-G+ DC-10 and sHLA-G plasma level were significantly higher in patients than healthy controls. | HLA-G might be a key immune suppressive molecule in tumors. | 2016 (36) |
| Gastric cancer | ELISA | sHLA-G plasma level was significantly higher in patients than healthy controls. | Analysis of sHLA-G plus CA125, CA19-9 or CA72-4 levels is potentially useful in tumor diagnosis. | 2016 (37) |
| Esophageal cancer | IHC ELISA | Cell surface HLA-G expression in tumor tissue was significantly higher than in normal adjacent tissue. sHLA-G plasma levels were significantly higher in patients than healthy controls. HLA-G overexpression was related to high IL-10 levels. Increased HLA-G expression in tumor tissue correlated with advanced grades and lymph node involvement, but there was no correlation between sHLA-G and clinical features. | HLA-G expression was associated with tumor progression and poor prognosis. | 2014 (30) |
| Pancreas cancer | IHC | HLA-G expression was correlated with tumor stage. Diffuse HLA-G expression was related to less tumor infiltration by immune cells and lower survival rate. | HLA-G expression was associated with tumor progression and poor prognosis. | 2015 (29) |
| Breast cancer | ELISA | Total and free sHLA-G levels in patients were significantly higher than in normal controls. sHLA-G in extracellular vesicles correlated with tumor progression, but free sHLA-G levels improved the clinical outcome. | For clinical outcome, evaluation of T cell subtypes is necessary. | 2016 (38) |
| Colorectal cancer | IHC | 70.7% of patients were HLA-G positive (expression above 5% was considered positive). High HLA-G expression (positive cells above 55%) was associated with advanced stages and lower survival than lower HLA-G expression. | High HLA-G expression was associated with poor prognosis. | 2017 (39) |
| Colorectal cancer | IHC ELISA | Levels of sHLA-G were higher in patients with mucinous carcinoma than other subtypes of colorectal cancer. In colorectal cancer, sHLA-G plasma levels were associated with shorter liver metastasis-free survival in stage II and with longer liver metastasis-free survival in stage III. High HLA-G expression in tumor tissues was related to low survival. | HLA-G is a potential surrogate marker for the prognosis of colorectal and sequential disease. | 2017 (40) |
| Colorectal cancer | ELISA | sHLA-G levels were significantly higher in patients with colorectal cancer than normal controls, and correlated with low survival and more advanced stages. sHLA-G in patients who died were obviously higher than in those who survived. | HLA-G expression was associated with tumor progression and poor prognosis. | 2017 (41) |
| Rectal cancer | IHC | High HLA-G and MHC I expression and Foxp3+ infiltrating cells above the median were related to better prognosis. | This pattern of immune marker expression creates a tumor phenotype related to better outcome, and could be used as a clinical prognostic marker for rectal cancer. | 2016 (42) |
FCM, flow cytometry; ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry
HLA-G displays different faces in hematological malignancies. In many studies, increased HLA-G expression was shown to be associated with a worse prognosis [49, 50], although high plasma levels of sHLA-G showed no correlation with prognosis [51, 52]. In contrast, several studies found evidence of a relationship between HLA-G and better prognosis for B cell neoplasms [53, 54]. These different observations can be attributed to the capacity of B cells to express ILT2 and therefore inhibit the proliferation of malignant B cells [55]. The fate of HLA-G interaction with its receptor on malignant B cells is determined by the balance between HLA-G-driven inhibition on these cells (as immune effector cells) and the antiproliferative effect on these cells (as target malignant cells) [55]. Heterogeneous patterns of HLA-G expression in different tumors may reflect differences in the biology of different types of tumor, genetic diversity among the populations studied to date, and the sensitivity of methods used to detect HLA-G [56]. In addition, the tumor microenvironment and viral infections may also have some impact on HLA-G expression [57], as discussed later in this review.
Associations between increased sHLA‑G plasma levels and poor prognosis for tumoral diseases have been reported in patients with HCC, lung cancer and colorectal cancer [2, 34, 35]. However, no clear association was observed between sHLA‑G plasma levels and clinicopathological features in breast cancer or gastric cancer [38, 58].
The evaluation of HLA-G might be used as a diagnostic tool to distinguish between malignant and benign tumors [31]. In addition, HLA-G might serve as a prognostic marker for the clinical outcome [36, 41]. HLA-G may also be a potential target for cancer immunotherapy in combination with other therapeutic strategies.
HLA-G promotes cancer development by different mechanisms. This molecule is involved in tumor immune escape not only by inhibiting NK cells but also by inducing apoptosis in NK cells and CTLs [27, 59]. Moreover, in many tumors HLA-G expression was reported to be positively associated with local Treg numbers [44, 45]. Soluble HLA-G, on the other hand, is able to downregulate chemokine receptors on lymphocytes and inhibit the chemotactic migration of NK, T and B cells into the tumor site [60, 61, 62]. As previously noted, HLA-G and HLA-E might cooperate in immune inhibition. Zeestraten et al. have shown that the absence of HLA-E and HLA-G on colon tumor cell surfaces was related to longer overall and disease-free survival [63]. Another study of colorectal cancer found that single or double expression of HLA-G and HLA-E was associated with shorter overall survival. In addition, HLA-E expression correlated significantly with tumor metastases. Another study reported that HLA-G expression alone can be used as a prognostic marker for overall survival [64]. HLA-G may also be involved in aggressive behavior of tumors and metastasis through the upregulation of matrix metalloproteinase 15 [65, 66]. Another mechanism called trogocytosis, i.e. the transfer of partial membranes with associated molecules via cell-cell contact [67], sometimes occurs between HLA-G-expressing tumor cells and NK or T cells, which makes the latter act as suppressor cells [68, 69, 70].
It was recently demonstrated that tumor cells secrete HLA-G-bearing extracellular vesicles. Riteau et al. first explained the presence of HLA-G-bearing extracellular vesicles in the supernatant of an HLA-G-positive melanoma cell line [71]. In this connection, König et al. and Grange et al. showed that these vesicles could modulate immune cell functions in kidney and breast cancers in ways that lead to cancer progression [38, 72].
HLA-G expression can be affected after cancer progression. Genetic instability and epigenetic changes due to progression may contribute to the development of less immunogenic tumors with increased HLA-G and decreased classical HLA class I molecules [56]. Furthermore, chronic inflammation and hypoxia in advanced tumors seem to induce HLA-G expression [73, 74]. HLA-G also may play a critical role in the creation of appropriate conditions for tumor progression, by generating tolerogenic APCs and Treg cells [75]. Upregulation of HLA-G might be induced by these regulatory/tolergenic cells, which produce IL-10 and TGF-β in the tumor microenvironment. This positive loop usually occurs in patients with advanced-stage cancer [76, 77].
HLA-G as a target in cancer therapy
Targeting the HLA-G molecule is one of several promising approaches for cancer therapy. In 2011, Agaugué et al. reported improved tumor lysis with a blocking monoclonal antibody against HLA-G in C57BL/6 mice which had been inoculated with the M8-HLA-G1 cell line [78]. The effect of blocking antibodies against HLA-G was also investigated by Maki et al., who observed that HLA-G blockade increased the susceptibility of chronic lymphocytic leukemia to NK cell-mediated killing [79]. Favier et al. also demonstrated that blocking ILT2 or HLA-G with a monoclonal antibody restored NK cell cytolytic activity on LCL 721.221 cells (a B lymphoblastoid cell line) which expressed HLA-G through transfection with HLA-G1 cDNA [80]. In another study, bone marrow-derived mesenchymal stem cells (MSCs) were treated with IL-10 to increase the secretion of HLA-G5. Neutralization of sHLA-G with an antibody suppressed the formation of CD4+/CD25high/Foxp3+ Treg cells and facilitated allogeneic T cell proliferation [81].
Zeng et al. showed that using HLA-G siRNA to decrease HLA-G expression on a human HCC cell line led to increased NK cell-induced tumor cytolysis [82]. This was despite the fact that HLA-G/ILT2 interaction can suppress the proliferation of Raji cells, a B cell line in Burkitt’s lymphoma. Therefore, in contrast to solid tumors, the administration of an HLA-G blocking antibody or HLA-G siRNA may lead to enhanced proliferation of malignant lymphocytes [54]. Our group recently reported improved NK cell cytotoxicity against SKOV3 cells after HLA-G downregulation with siRNA [83].
HLA-G and infection-related cancers
In infectious diseases, HLA-G has been shown to have deleterious effects by promoting pathogen escape from immune recognition [84]. Although it was found that oncoviruses directly induce cancer, some nononcogenic pathogens may also lead to malignancy by inducing chronic inflammation [85]. The mechanism of tumorigenesis by infectious agents has not been fully elucidated; however, it is clear that both host and pathogen are involved in this process [86]. In this regard, the main question is whether HLA-G plays role in tumorigenesis following infections.
Helicobacter pylori has been known as a dominant species living in the human stomach, where colonization by this pathogen sometimes leads to chronic inflammation, called gastritis. Although H. pylori is considered a strong risk factor for gastric cancers, only a small number of infected individuals develop malignancy. The risk of malignancy is highly related to both bacterial strain and host responses [87]. The only study to explore the relationship between HLA-G and H. pylori was that by Souza et al., who observed HLA-G expression in gastric tissues from 79.6% of patients with H. pylori infection. They found a negative association between the presence of HLA-G and colonization by H. pylori; however, the role of HLA-G in the induction of gastric cancer was not clarified in this study [88]. Recently, our group explored HLA-G expression in patients with H. pylori infection and gastric cancer. Our results showed that 32% of gastric cancer samples were positive for H. pylori, whereas only 12% of the samples were positive for both H. pylori and HLA-G (unpublished data).
Hepatitis B virus (HBV) is a known risk factor for HCC [86]. Park et al. detected higher levels of sHLA-G in patients with active HBV and HCC than healthy controls. Their results also showed increased levels of sHLA-G in early stages of HBV-mediated HCC [89]. However, in a recent study, we observed no correlation between HLA-G expression and the presence of HBV genome in HCC tissues [48]. Dong et al. observed a trend toward increasing HLA-G expression with tumor progression in patients with cervical intraepithelial neoplasia. They also detected higher expression of HLA-G in human papillomavirus (HPV)-positive patients than HPV-negative patients with this disease [90]. Guimarães et al. reported low HLA-G5 levels in their patients with HPV-related cervical cancers. They also observed lower HLA-G5 levels in invasive cervical cancers without metastases compared to patients with metastases [91]. In recent work by our group on gynecologic cancers, higher levels of HLA-G expression were detected in cervical cancer than other gynecologic cancers, and HPV-DNA was detected in 65% of these lesions (unpublished data).
HLA-G expression may be controlled more directly by local microenvironmental factors than by infectious agents. This hypothesis was supported by Gazit et al., who observed HLA-G induction by culture conditions such as nutrient deficiency, hypoxia, or both in an EBV-transformed B cell line [92].
HLA-G: regulation of gene expression
In addition to epigenetic factors, HLA-G expression is controlled by one or more inherited alleles and genetic variations in noncoding regions of the gene [93, 94]. Therefore, the expression level of HLA-G after infections appears to be determined, at least in part, by the host’s genetic background.
Low expression of HLA-G due to promoter hypermethylation was reported to be associated with preeclampsia [95]. Although HLA-G overexpression has been documented in several cancers, the role of HLA-G promoter demethylation in these processes has not been clarified. Gillio-Tos et al. found no relation between HLA-G promoter demethylation and HPV-induced cervical intraepithelial neoplasia [96].
It was believed that HLA-G expression might also be affected by polymorphisms in the 5′ upstream regulatory region (5′URR), because transcription factor binding sites are mostly located in this region, e.g. cyclic AMP-dependent transcription factor-1 (ATF-1), Ras responsive element-binding protein1 (RREB1), interferon regulatory factor-1 (IRF-1) and nuclear factor (NF)-κB [97].
HLA-G expression is known to be induced by local changes resembling hypoxia, or by the presence of progesterone. Hypoxia response element is located in the upstream region of the HLA-G gene, at positions −242 to −238 [98]. The progesterone receptor, after binding to its ligand, is also bound to progesterone response element in the HLA-G promoter at positions −52 and −38 [99]. It has also been reported that HLA-G expression can be upregulated by IL-10, cortisol and some drugs such as methotrexate, albeit via unknown pathways [77, 100, 101].
In addition to factors involved at the transcriptional level, HLA-G expression is also controlled at post-transcriptional levels [102]. Although some microRNAs bind to nonpolymorphic sequences, polymorphisms in the HLA-G 3' untranslated region (3'UTR) may affect mRNA stability by changing the affinity of this region for the corresponding micoRNAs [94, 102]. Moreover, HLA-G expression is also affected by the level of related microRNAs. Bian et al. have shown that decreased miR-152 expression leads to HLA-G upregulation and impaired NK cell-mediated antitumor response [103]. Jasinski-Bergner et al. also reported that miR-548q and miR-628-5p influenced HLA-G downregulation and enhanced NK cell-mediated cytolysis [104]. One of the most frequently studied polymorphisms in 3'UTR is the 14-bp insertion/deletion (ins/del). A link between the 14-bp del/del genotype and higher levels of HLA-G production was reported in healthy people [105].
A meta-analysis by Zhang et al. explored the role of HLA-G 14-bp ins/del polymorphism in the susceptibility to cervical cancer, HCC, esophageal cancer, neuroblastoma and papillary thyroid carcinoma. They found a prominent role for this polymorphism in susceptibility to HCC but not to the other cancers they reviewed [106]. However, the association between this polymorphism and cancer susceptibility may differ among populations [97]. For example, the frequency of the HLA-G 14-bp del/del genotype was higher in patients from southeastern Iran who had breast cancer compared to the control population [107], whereas this genotype showed no association with breast cancer in patients from northwestern Iran [108]. Therefore, HLA‑G polymorphisms might be predictive markers for susceptibility to some types of cancer. However, additional studies of populations representing different ethnicities and with larger sample sizes are required to confirm this hypothesis.in Conclusion, The extensive immunomodulatory effects of HLA-G on malignancies have opened a new window for the diagnosis, prognosis and treatment of cancers. HLA-G is a potential new biomarker for the differential diagnosis between malignant and benign tumors. At this time, however, HLA-G is a potential target molecule in therapeutic approaches to enhance the efficacy of cancer treatments. Therefore, more in vitro and in vivo studies are needed to determine the therapeutic potential HLA-G molecules.
Acknowledgments
This work was supported by Shiraz University of Medical Sciences. We thank K. Shashok (AuthorAID in the Eastern Mediterranean) for improving the use of English in the manuscript.
References
[1]. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68(3):369-95.
[2]. Curigliano G, Criscitiello C, Gelao L, Goldhirsch A. Molecular pathways: human leukocyte antigen G (HLA-G). Clin Cancer Res. 2013;19(20):5564-71.
[3]. Alegre E, Rizzo R, Bortolotti D, Fernandez-Landázuri S, Fainardi E, González A. Some basic aspects of HLA-G biology. Journal of immunology research.2014.
[4]. Baudhuin J, Lesport E, Sousa S, Migraine J, Vigneron J, Lemaoult J, et al. HLA‐G inhibition of NK‐cell cytolytic function is uncoupled from tumor cell lipid raft reorganization. European journal of immunology. 2012;42(3):700-9.
[5]. Fons P, Chabot S, Cartwright JE, Lenfant F, L'Faqihi F, Giustiniani J, et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood. 2006;108(8):2608-15.
[6]. Rouas-Freiss N, Moreau P, Menier C, LeMaoult J, Carosella ED. Expression of tolerogenic HLA-G molecules in cancer prevents antitumor responses. Semin Cancer Biol. 2007;17(6):413-21.
[7]. Fournel S, Aguerre-Girr M, Huc X, Lenfant F, Alam A, Toubert A, et al. Cutting edge: soluble HLA-G1 triggers CD95/CD95 ligand-mediated apoptosis in activated CD8+ cells by interacting with CD8. J Immunol. 2000;164(12):6100-4.
[8]. LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005;19(6):662-4.
[9]. Amodio G, Sales de Albuquerque R, Gregori S. New insights into HLA‐G mediated tolerance. HLA. 2014;84(3):255-63.
[10]. LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci U S A. 2004;101(18):7064-9.
[11]. Morandi F, Pistoia V. Interactions between HLA-G and HLA-E in physiological and pathological conditions. Frontiers in immunology. 2014;5:394.
[12]. Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proceedings of the national academy of sciences of the United States of America. 1987;84(24):9145-9.
[13]. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248(4952):220-3.
[14]. Comiskey M, Warner CM, Schust DJ. MHC molecules of the preimplantation embryo and trophoblast. 2013.
[15]. Ferreira LM, Meissner TB, Tilburgs T, Strominger JL. HLA-G: At the Interface of Maternal–Fetal Tolerance. Trends in immunology. 2017;38(4):272-86.
[16]. Rouas-Freiss N, Kirszenbaum M, Dausset J, Carosella ED. [Fetomaternal tolerance: role of HLA-G molecule in the protection of the fetus against maternal natural killer activity]. C R Acad Sci III. 1997;320(5):385-92.
[17]. Amodio G, Mugione A, Sanchez AM, Vigano P, Candiani M, Somigliana E, et al. HLA-G expressing DC-10 and CD4(+) T cells accumulate in human decidua during pregnancy. Hum Immunol. 2013;74(4):406-11.
[18]. Gregori S, Amodio G, Quattrone F, Panina-Bordignon P. HLA-G Orchestrates the Early Interaction of Human Trophoblasts with the Maternal Niche. Front Immunol. 2015;6:128.
[19]. Gonen-Gross T, Goldman-Wohl D, Huppertz B, Lankry D, Greenfield C, Natanson-Yaron S, et al. Inhibitory NK receptor recognition of HLA-G: regulation by contact residues and by cell specific expression at the fetal-maternal interface. PLoS One. 2010;5(1):e8941
[20]. Gonen-Gross T, Achdout H, Arnon TI, Gazit R, Stern N, Hořejší V, et al. The CD85J/leukocyte inhibitory receptor-1 distinguishes between conformed and β2-microglobulin-free HLA-G molecules. The journal of immunology. 2005;175(8):4866-74.
[21]. Li C, Houser BL, Nicotra ML, Strominger JL. HLA-G homodimer-induced cytokine secretion through HLA-G receptors on human decidual macrophages and natural killer cells. Proceedings of the national academy of sciences. 2009;106(14):5767-72.
[22]. Brugiere O, Thabut G, Krawice‐Radanne I, Rizzo R, Dauriat G, Danel C, et al. Role of HLA‐G as a Predictive Marker of Low Risk of Chronic Rejection in Lung Transplant Recipients: A Clinical Prospective Study. American journal of transplantation. 2015;15(2):461-71.
[23]. Okushi Y, Okino K, Mukai K, Matsui Y, Hayashi N, Fujimoto K, et al. Circulating and renal expression of HLA-G prevented chronic renal allograft dysfunction in Japanese recipients. Clin Exp Nephrol. 2017;21(5):932-40.
[24]. Dunn GP, Fecci PE, Curry WT. Cancer immunoediting in malignant glioma. Neurosurgery. 2012;71(2):201-22; discussion 22-3.
[25]. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991-8.
[26]. Lin A, Yan WH, Xu HH, Gan MF, Cai JF, Zhu M, et al. HLA-G expression in human ovarian carcinoma counteracts NK cell function. Ann Oncol. 2007;18(11):1804-9.
[27]. Lesport E, Baudhuin J, LeMaoult J, Sousa S, Doliger C, Carosella ED, et al. Human melanoma cell secreting human leukocyte antigen–G5 inhibit natural killer cell cytotoxicity by impairing lytic granules polarization toward target cell. Human immunology. 2009;70(12):1000-5.
[28]. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci U S A. 1998;95(8):4510-5.
[29]. Zhou L, Niu ZY, Liang ZY, Zhou WX, You L, Wang MY, et al. HLA-G impairs host immune response and predicts poor prognosis in pancreatic cancer. Am J Transl Res. 2015;7(10):2036-44.
[30]. Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G, et al. Human leukocyte antigen G is associated with esophageal squamous cell carcinoma progression and poor prognosis. Immunology letters. 2014;161(1):13-9.
[31]. Provatopoulou X, Kalogera E, Sagkriotis A, Zagouri F, Nonni A, Zografos GC, et al. Soluble human leukocyte antigen-G expression in patients with ductal and lobular breast malignancy. Anticancer Res. 2012;32(3):1021-6.
[32]. Menier C, Rouas-Freiss N, Carosella ED. The HLA-G non classical MHC class I molecule is expressed in cancer with poor prognosis. Implications in tumour escape from immune system and clinical applications. 2009.
[33]. Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human leukocyte antigen-G (HLA-G) polymorphism and expression in breast cancer patients. PLoS One. 2014;9(5):e98284.
[34]. Lin A, Zhu CC, Chen HX, Chen BF, Zhang X, Zhang JG, et al. Clinical relevance and functional implications for human leucocyte antigen-g expression in non-small-cell lung cancer. J Cell Mol Med. 2010;14(9):2318-29.
[35]. Wang Y, Ye Z, Meng XQ, Zheng SS. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2011;10(2):158-63.
[36]. Xu DP, Shi WW, Zhang TT, Lv HY, Li JB, Lin A, et al. Elevation of HLA-G-expressing DC-10 cells in patients with gastric cancer. Hum Immunol. 2016;77(9):800-4.
[37]. Pan YQ, Ruan YY, Peng JB, Han QY, Zhang X, Lin A, et al. Diagnostic significance of soluble human leukocyte antigen-G for gastric cancer. Hum Immunol. 2016;77(4):317-24.
[38]. Konig L, Kasimir-Bauer S, Hoffmann O, Bittner AK, Wagner B, Manvailer LF, et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum Immunol. 2016;77(9):791-9.
[39]. Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget. 2017;8(64):107441-51.
[40]. Kirana C, Ruszkiewicz A, Stubbs RS, Hardingham JE, Hewett PJ, Maddern GJ, et al. Soluble HLA‐G is a differential prognostic marker in sequential colorectal cancer disease stages. International journal of cancer. 2017;140(11):2577-86.
[41]. Li JB, Ruan YY, Hu B, Dong SS, Bi TN, Lin A, et al. Importance of the plasma soluble HLA-G levels for prognostic stratification with traditional prognosticators in colorectal cancer. Oncotarget. 2017;8(30):48854-62.
[42]. Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, et al. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer. 2014;14:486.
[43]. Gonzalez A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The immunosuppressive molecule HLA-G and its clinical implications. Crit Rev Clin Lab Sci. 2012;49(3):63-84.
[44]. Du L, Xiao X, Wang C, Zhang X, Zheng N, Wang L, et al. Human leukocyte antigen‐G is closely associated with tumor immune escape in gastric cancer by increasing local regulatory T cells. Cancer science. 2011;102(7):1272-80.
[45]. Tuncel T, Karagoz B, Haholu A, Ozgun A, Emirzeoglu L, Bilgi O, et al. Immunoregulatory function of HLA-G in gastric cancer. Asian Pac J Cancer Prev. 2013;14(12):7681-4.
[46]. Gan L-H, Huang L-F, Zhang X, Lin A, Xu D-P, Wang Q, et al. Tumor-specific upregulation of human leukocyte antigen–G expression in bladder transitional cell carcinoma. Human immunology. 2010;71(9):899-904.
[47]. Locafaro G, Amodio G, Tomasoni D, Tresoldi C, Ciceri F, Gregori S. HLA-G expression on blasts and tolerogenic cells in patients affected by acute myeloid leukemia. J Immunol Res. 2014;2014:636292.
[48]. Khodabandeh Shahraki P, Zare Y, Azarpira N, Hosseinzadeh M, Farjadian S. Prognostic Value of HLA-G in Malignant Liver and Pancreas Lesions. Iran J Immunol. 2018;15(1):28-37.
[49]. Alkhouly N, Shehata I, Ahmed MB, Shehata H, Hassan S, Ibrahim T. HLA-G expression in acute lymphoblastic leukemia: a significant prognostic tumor biomarker. Med Oncol. 2013;30(1):460.
[50]. Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, et al. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia. 2008;22(5):998-1006.
[51]. Amiot L, Le Friec G, Sebti Y, Drenou B, Pangault C, Guilloux V, et al. HLA-G and lymphoproliferative disorders. Semin Cancer Biol. 2003;13(5):379-85.
[52]. Perez-Chacon G, Rosado S, Rebolleda N, Losada-Fernandez I, Vargas JA, Morado M, et al. Prognostic irrelevance of HLA-G in B-cell chronic lymphocytic leukemia. Int J Lab Hematol. 2009;31(3):327-37.
[53]. Jesionek-Kupnicka D, Bojo M, Prochorec-Sobieszek M, Szumera-Cieckiewicz A, Jablonska J, Kalinka-Warzocha E, et al. HLA-G and MHC Class II Protein Expression in Diffuse Large B-Cell Lymphoma. Arch Immunol Ther Exp (Warsz). 2016;64(3):225-40.
[54]. Naji A, Menier C, Maki G, Carosella E, Rouas-Freiss N. Neoplastic B-cell growth is impaired by HLA-G/ILT2 interaction. Leukemia. 2012;26(8):1889.
[55]. Carosella ED, Rebmann V. A controversy on HLA-G and B-cell malignancies? Journal of Immunotherapy applications. 2016;3(1):1.
[56]. Urosevic M, Dummer R. Human leukocyte antigen–G and cancer immunoediting. Cancer research. 2008;68(3):627-30.
[57]. Oucherif O, Naimi D. Function of HLA-G in cancer immunoediting and its clinical benefitsFonction de HLA-G dans le cancer immunoediting et ses avantages cliniques. J Africain du Cancer/African Journal of Cancer. 2015;7(3):132-9.
[58]. Chen H-X, Lin A, Shen C-J, Zhen R, Chen B-G, Zhang X, et al. Upregulation of human leukocyte antigen–G expression and its clinical significance in ductal breast cancer. Human immunology. 2010;71(9):892-8.
[59]. Contini P, Ghio M, Poggi A, Filaci G, Indiveri F, Ferrone S, et al. Soluble HLA‐A,‐B,‐C and‐G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. European journal of immunology. 2003;33(1):125-34.
[60]. Naji A, Menier C, Morandi F, Agaugue S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol. 2014;192(4):1536-46.
[61]. Morandi F, Ferretti E, Bocca P, Prigione I, Raffaghello L, Pistoia V. A novel mechanism of soluble HLA-G mediated immune modulation: downregulation of T cell chemokine receptor expression and impairment of chemotaxis. PLoS One. 2010;5(7):e11763.
[62]. Morandi F, Ferretti E, Castriconi R, Dondero A, Petretto A, Bottino C, et al. Soluble HLA-G dampens CD94/NKG2A expression and function and differentially modulates chemotaxis and cytokine and chemokine secretion in CD56bright and CD56dim NK cells. Blood. 2011;118(22):5840-50.
[63]. Zeestraten E, Reimers M, Saadatmand S, Dekker JT, Liefers G, Van Den Elsen P, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. British journal of cancer. 2014;110(2):459.
[64]. Guo ZY, Lv YG, Wang L, Shi SJ, Yang F, Zheng GX, et al. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol. 2015;293(1):10-6.
[65]. Liu X, Gu W, Li X. HLA-G regulates the invasive properties of JEG-3 choriocarcinoma cells by controlling STAT3 activation. Placenta. 2013;34(11):1044-52.
[66]. Lin A, Xu HH, Xu DP, Zhang X, Wang Q, Yan WH. Multiple steps of HLA-G in ovarian carcinoma metastasis: alter NK cytotoxicity and induce matrix metalloproteinase-15 (MMP-15) expression. Hum Immunol. 2013;74(4):439-46.
[67]. Carosella ED, Favier B, Rouas-Freiss N, Moreau P, Lemaoult J. Beyond the increasing complexity of the immunomodulatory HLA-G molecule. Blood. 2008;111(10):4862-70.
[68]. Caumartin J, Favier B, Daouya M, Guillard C, Moreau P, Carosella ED, et al. Trogocytosis-based generation of suppressive NK cells. EMBO J. 2007;26(5):1423-33.
[69]. LeMaoult J, Caumartin J, Daouya M, Favier B, Le Rond S, Gonzalez A, et al. Immune regulation by pretenders: cell-to-cell transfers of HLA-G make effector T cells act as regulatory cells. Blood. 2007;109(5):2040-8.
[70]. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood. 2012;120(10):2055-63.
[71]. Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, et al. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. 2003;64(11):1064-72.
[72]. Grange C, Tapparo M, Tritta S, Deregibus MC, Battaglia A, Gontero P, et al. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer. 2015;15:1009.
[73]. Zidi I, Guillard C, Marcou C, Krawice-Radanne I, Sangrouber D, Rouas-Freiss N, et al. Increase in HLA-G1 proteolytic shedding by tumor cells: a regulatory pathway controlled by NF-κB inducers. Cellular and molecular life sciences CMLS. 2006;63(22):2669-81.
[74]. Mouillot G, Marcou C, Zidi I, Guillard C, Sangrouber D, Carosella ED, et al. Hypoxia modulates HLA-G gene expression in tumor cells. Hum Immunol. 2007;68(4):277-85.
[75]. Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends Immunol. 2008;29(3):125-32.
[76]. Guan Z, Song B, Liu F, Sun D, Wang K, Qu H. TGF-β induces HLA-G expression through inhibiting miR-152 in gastric cancer cells. Journal of biomedical science. 2015;22(1):107.
[77]. Rodríguez JA, Galeano L, Palacios DM, Gómez C, Serrano ML, Bravo MM, et al. Altered HLA class I and HLA-G expression is associated with IL-10 expression in patients with cervical cancer. Pathobiology. 2012;79(2):72-83.
[78]. Agaugué S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood. 2011;117(26):7021-31.
[79]. Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, et al. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia. 2008;22(5):998-1006.
[80]. Favier B, LeMaoult J, Lesport E, Carosella ED. ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. The FASEB Journal. 2010;24(3):689-99.
[81]. Selmani Z, Naji A, Zidi I, Favier B, Gaiffe E, Obert L, et al. Human leukocyte antigen‐G5 secretion by human mesenchymal stem cells is required to suppress T lymphocyte and natural killer function and to induce CD4+ CD25highFOXP3+ regulatory T cells. Stem cells. 2008;26(1):212-22.
[82]. Zeng XC, Zhang T, Huang DH, Wang GY, Chen W, Li H, et al. RNA interfering targeting human leukocyte antigen-G enhanced immune surveillance mediated by the natural killer cells on hepatocellular carcinoma. Ann Clin Lab Sci. 2013;43(2):135-44.
[83]. Nazari N, Farjadian S. Natural Killer Cell Cytotoxicity Against SKOV3 after HLA-G Downregulation by shRNA. Iran J Immunol. 2016;13(3):178-85.
[84]. Amiot L, Vu N, Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. 2014;2014:298569.
[85]. Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339(6117):286-91.
[86]. Zamor PJ, deLemos AS, Russo MW. Viral hepatitis and hepatocellular carcinoma: etiology and management. J Gastrointest Oncol. 2017;8(2):229-42.
[87]. Polk DB, Peek Jr RM. Helicobacter pylori: gastric cancer and beyond. Nature reviews cancer. 2010;10(6):403.
[88]. Souza DM, Genre J, Silva TG, Soares CP, Rocha KB, Oliveira CN, et al. Upregulation of Soluble HLA-G5 and HLA-G6 Isoforms in the Milder Histopathological Stages of Helicobacter pylori Infection: A Role for Subverting Immune Responses? Scand J Immunol. 2016;83(1):38-43.
[89]. Park Y, Park Y, Lim HS, Kim YS, Hong DJ, Kim HS. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens. 2012;79(2):97-103.
[90]. Dong D-d, Yang H, Li K, Xu G, Song L-h, Fan X-l, et al. Human leukocyte antigen-G (HLA-G) expression in cervical lesions: association with cancer progression, HPV 16/18 infection, and host imhypomune response. Reproductive sciences. 2010;17(8):718-23.
[91]. Guimarães MC, Soares CP, Donadi EA, Derchain SF, Andrade LA, Silva TG, et al. Low expression of human histocompatibility soluble leukocyte antigen-G (HLA-G5) in invasive cervical cancer with and without metastasis, associated with papilloma virus (HPV). Journal of histochemistry and cytochemistry. 2010;58(5):405-11.
[92]. Gazit E, Sherf M, Balbin E, Muratov A, Goldstein I, Loewenthal R. HLA-G expression is induced in Epstein-Barr virus–transformed B-cell lines by culture conditions. Human immunology. 2007;68(6):463-8.
[93]. Castelli EC, Mendes-Junior CT, Veiga-Castelli LC, Roger M, Moreau P, Donadi EA. A comprehensive study of polymorphic sites along the HLA-G gene: implication for gene regulation and evolution. Mol Biol Evol. 2011;28(11):3069-86.
[94]. Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, et al. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. 2017;19:118-27.
[95]. Tang Y, Liu H, Li H, Peng T, Gu W, Li X. Hypermethylation of the HLA-G promoter is associated with preeclampsia. Molecular human reproduction. 2015;21(9):736-44.
[96]. Gillio-Tos A, Bicalho Mda G, Fiano V, Grasso C, Tarallo V, De Marco L, et al .Case-control study of HLA- promoter methylation status, HPV infection and cervical neoplasiain Curitiba, Brazil: a pilot analysis. BioMed Central cancer. 2012;12:618.
[97]. Dias FC, Castelli EC, Collares CV, Moreau P, Donadi EA. The Role of HLA-G Molecule and HLA-G Gene Polymorphisms in Tumors, Viral Hepatitis, and Parasitic Diseases. Front Immunol. 2015;6:9.
[98]. Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005(306):re12-re.
[99]. Yie S-m, Xiao R, Librach CL. Progesterone regulates HLA-G gene expression through a novel progesterone response element. Human Reproduction. 2006;21(10):2538-44.
[100]. Moreau P, Faure O, Lefebvre S, Ibrahim EC, O'Brien M, Gourand L, et al. Glucocorticoid hormones upregulate levels of HLA-G transcripts in trophoblasts. Transplant Proc. 2001;33(3):2277-80.
[101]. Rizzo R, Rubini M, Govoni M, Padovan M, Melchiorri L, Stignani M, et al. HLA-G 14-bp polymorphism regulates the methotrexate response in rheumatoid arthritis. Pharmacogenetics and genomics. 2006;16(9):615-23.
[102]. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. 2014;2014:734068.
[103]. Bian X, Si Y, Zhang M, Wei R, Yang X, Ren H, et al. Down-expression of miR-152 lead to impaired anti-tumor effect of NK via upregulation of HLA-G. Tumour Biol. 2016;37(3):3749-56.
[104]. Jasinski-Bergner S, Reches A, Stoehr C, Massa C, Gonschorek E, Huettelmaier S, et al. Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget. 2016;7(18):26866.
[105]. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3’untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PloS one. 2013;8(10):e71742.
[106]. Zhang S, Wang HT. Association between HLA-G 14-bp insertion/deletion polymorphism and cancer risk: a meta-analysis. Official Journal of the Balkan Union of Oncology. 2014; 19(2):567-72.
[107]. Eskandari-Nasab E, Hashemi M, Hasani SS, Omrani M, Taheri M, Mashhadi MA. Association between HLA-G 3'UTR 14-bp ins/del polymorphism and susceptibility to breast cancer. Cancer Biomark. 2013;13(4):253-9.
[108]. Haghi M, Hosseinpour Feizi MA, Sadeghizadeh M, Lotfi AS. 14-bp Insertion/Deletion Polymorphism of the HLA-G gene in Breast Cancer among Women from North Western Iran. Asian Pacific journal of cancer prevention. 2015;16(14):6155-8.
License
Copyright
© ,
Author Details