Detection of miR-21, MUC1mRNA and VEGF Protein Biomarkers Expression Changes in Oral Squamous Cell Carcinomas (OSCC) in Peripheral Blood
Download
Abstract
Background and purpose: Oral squamous cell carcinoma is a common type of cancer and one of the most important causes of death in the world; therefore, this study investigates the expression of miR-21, MUC1 mRNA and VEGF protein in peripheral blood for early detection and better treatment of this cancer using Real-Time RT-PCR and Elisa.
Materials and methods: In this experimental study, 50 OSCC patients and 50 healthy subjects were selected as controls. The total RNA in peripheral blood was extracted by RNA extraction kit and quality evaluation of extracted RNA was measured by NanoDrop.For the synthesis of cDNA, Viva 2-steps RT-PCR kit was used. Expression of miR-21 and MUC1 mRNA were measured by Real-Time RT-PCR and used Elisa for VEGF protein. The relationship between the expressions of these biomarkers was evaluated with tumor staging and progression of cancer.
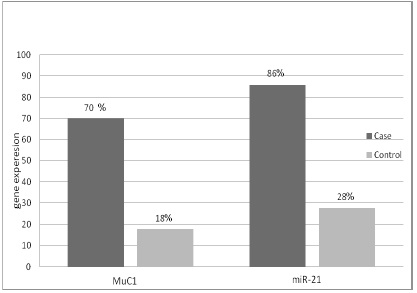
Results: The average age difference is not significant in these two groups by using t-test. The miR-21 was positive in 43 out of 50 OSCC patients (86%), and it was also 14 out of 50 healthy subjects (28%). The MUC1 mRNA was positive in 35 out of 50 OSCC patients, indicating 70% sensitivity, and it was positive in 9 of the 50 healthy subjects(18%). The serum levels of VEGF protein were measured using Elisa; it was positive in 32 out of 50 OSCC patients (64%) and this biomarker in healthy subjects report was not positive, indicating that the specificity is 100%.
Conclusions: Overall, the miR-21, MUC1 mRNA and VEGF protein biomarkers could be suggested as a diagnostic screening test for the early detection of diseases. To prove further research is recommended further studies with a larger sample size to be done.
Introduction
One of the most frequent cancers in the world is Oral Squamous Cell Carcinoma (OSCC) and consists about one to three percent of all human cancers[1 2, ]. unfortunately, due to its poor prognosis, this kind of cancer continues to show a poor prognosis and remains a lethal disease for more than fifty percent of cases diagnosed annually[2 3, ]. Oral cavity cancer is an important problem in the world with nearly 263,900 new cases and 128,000 deaths worldwide in 2008, and oral squamous cell carcinoma almost includes 90% of all oral malignancies[1 4, ]. Despite advances in clinical research in the past 30 years, the survival rate of Oral squamous cell carcinoma patients has not improved[5]. Lymph node metastasis and prognostic factors for survival of Oral squamous cell carcinoma patients, but are usually diagnosed in time for lies[6 7, ].
Additionally, patients treated with surgery often results poorly due to unexpected events such as regional recurrence[6 8, ]. Therefore, prognostic factors can be useful in helping to guide treatment decisions. However, useful prognostic markers and predictive assays for clinical diagnosis and treatment of Oral squamous cell carcinoma have been established[9].In the present study3, prognostic markers for early diagnosis of Oral squamous cell carcinoma are studied.
Mucins
To date, 21 mucins have been identified in the human body and according to the type of operation are divided into membrane-bound and secreted types. These mucins play a major role in cell signaling, cell growth, cell adhesion, immune response, differentiation and help to protect and lubricate luminal surfaces under normal physiological conditions[10].In the present study, mucin expression level in Oral squamous cell carcinoma patients was examined and it was found that the expression of these markers in these patients increased. In fact, this marker is associated with tumor size and tumor staging.
MicroRNAs
MicroRNAs (miRNAs) are a family of small noncoding RNAs of 18-25 nucleotides in length, which have a critical role in the posttranscriptional regulation of gene expression through base-pair binding to the 3′-untranslated region (3′-UTR) of their target mRNAs[11]. The microRNAs play a role in biological and pathological processes, including the cell cycle, metabolism, differentiation, aging, development, patterning, apoptosis, death, etc[12]. Some microRNAs have been functionally classified as tumor suppressors or proto-oncogenes[13 14, ]. One of the most intensively studied microRNAs is miR-21 (miRNA-21). MiR-21 expression pattern may change in different disease states[15]. MiR-21 has been identified as the microRNAs that is overexpressed in solid tumors, and its levels have been increased in very diverse cancer types including breast, glioblastoma, liver and pancreatic cancers[16 17, ].
VEGF protein
Vascular endothelial growth factor (VEGF) is a cytokine that causes sprouting of endothelial cells, cell proliferation, migration, disease progression, prognosis, tumor angiogenesis, and vascular permeability and is involved in every stage of vascular development[18 19, ].Therefore, VEGF is considered an important goal of therapeutic target. Studies have shown that the expression of VEGF associated with metastasis and tumor invasion pattern [20, 21, 22]. Now the relation of VEGF expression with pathologic differentiation and clinical stage are controversial. Moreover, The relation between tumor microvessel density (MVD) and the prognosis is still controversial[23 24, ]. The aim of this study is to investigate the relation between the expression of VEGF in oral squamous cell carcinoma and angiogenesis.
Materials and Methods
Patient samples
In the present study, fifty peripheral blood samples were collected from patients of matched OSCC at the Cancer Institute of Tehran Medicine University (Tehran, Iran), and fifty non-cancerouspatients’volunteers were used as references voluntarily. The minimum and maximum age of these persons is 22 and 77 years, respectively. An informed consent was obtained from all persons in this study.
Pre-processing of blood samples
From each person, 12 mL of peripheral blood were obtained by sampling syringe in glass test tubes. Then, 2mL of the sample was discarded and 10mL sample was divided into 8 mL and 2 mL portions. The 8 mL portion was added to the Falcon tube containing anticoagulant EDTA reagent, and the 2 mL portion was coagulated for serum extraction. The 2 mL portion sample was centrifuged at 3000rpm at room temperature, the serum samples were collected as 1 mL aliquots in polyethylene tubes by a sampler for the determination of VEGF, and RNA was extracted immediately from the 8 mL non-coagulated portion.
Extraction of RNA
For the extraction of Total RNA of peripheral blood from 8 mL portions, the standard kit of Qiagen RNeasy Midi Kit (Cat.No.75144; Qiagen, Germany) following the manufacturer’s instructions was used.
Efficiently and quality Evaluation of extracted RNA
For the quality evaluation of RNA, absorbance of extracted RNA was determined at the range of 260-280 nm by NanoDrop spectrophotometer(Bio-Tek, USA). Extracted RNA was immediately applied for reverse transcription.
cDNA synthesis
For the preparation of cDNA from extracted mRNA, Viva 2-steps RT-PCR kit (Cat. No. RTPL12; Vivantis, Malaysia), kept at -20ºC with its standard procedure, was used. The quality evaluation of cDNA was determined by NanoDrop spectrophotometer (Bio-Tek, USA).
Design of specific primers
For each of the biomarkers (miR-21and MUC1 mRNA), the specific primers were designed by AlleleID6 software (Premier Biosoft, Palo Alto, USA) and ordered for synthesis as the lyophilized powder. Then the 100 pm.µL-1 (picomole per microliter) the solution of primers was prepared by dissolving of appreciating the amount of powder in autoclaved deionized water (storage at -20ºC). For the final use of the primers, a 10 pm.µL-1working solution was prepared by dilution and used in the final reaction.
Real-Time RT-PCR
Real-Time RT-PCR reaction was performed using Titan HotTeq EvaGreen qPCR Mix(ROX) kit (Sinaclon, Iran) in a CFX96 qRT-PCR detection system (cat. No. BT11101; BioRed, USA). Reactions were carried out in a total of 20µl, including 4µl PCR Master Mix, 0.5 µl forward primer, 0.5 µl reverse primer, 2µl DNA template and deionized distilled water up to a final volume of 20 µl. All samples were processed in duplicate using HotTeq EvaGreen qPCR Mix with the following cycling condition in Tabl 1. The 5srRNA and 18srRNA genes were used as internal references for miR-21 and MUC1 mRNA biomarkers, respectively. For MUC1, the forward and reverse primers were GTGCCCCCTAGCAGTACCG and GACGTGCCCCTACAAGTTGG, respectively, and after the endpoint of reaction, the results were interpreted based on amplification and melting curves.
| Cycles | Time | Temperature | Cycle step |
| 1 | 15 min | C֯95 | Initial denaturation |
| 40 | 15 s | C ֯95 | Denaturation |
| 20 s | C ֯ 65֯60- | Annealing | |
| 20 s | C ֯72 | Elongation |
VEGF serum level determination
The VEGF serum levels were determined by Elisa method using Bioassay Technology Laboratory kit, at room temperature.
Statistical analysis
The differences in mean between miR-21, MUC1 mRNA, VEGF protein expression in the sample of oral squamous cell carcinoma and non-cancerous samples were analyzed using t-test. Statistical analysis was performed with the statistical software SPSS, version 20 (SPSS Inc., Chicago, IL, USA).
Results
Clinical- and demographic particulars of persons
To specificity, I explore the aging role in OSCC, t-test method was performed, and the results have been listed in Tabl 2. These two groups have the same average, and the results show that the age factor does not have any significant effect on OSCC.
| main group | age (year) | ||
| age range | mean | SD | |
| Patients (50 person) | 25-70 | 45.95 | 10.86 |
| Noncancerous (50 person) | 24-70 | 48.85 | 12.36 |
| SD: Standard Deviation | P-value = 0.442 |
Quantizing the expression of the study housekeeping genes
In this study, the housekeeping gene (reference gene)for marker miR-21 is 5srRNA and the reference gene for marker MUC1 mRNA is 18srRNA. The expression level of the reference gene (5srRNA, 18srRNA) was quantized for each sample. The mean value of this reference gene between the controls and OSCC patients of this study showed no significant difference (p=0.442); so, therefore this gene was selected as a reference gene.
Real-Time PCR
Two different methods can be used for performing Real-Time PCR, (1) absolute quantification and (2) relative quantification. In this work, the relative quantification was used. Two prepared cDNA vial from cancerous and non-cancerous persons were tested for reference gene and biomarkers, and the results were obtained by the melting curve method.
Expression analyses of markers
We investigated the expression levels of MUC1 mRNA and miR-21, in peripheral blood samples of oral squamous cell carcinoma (n=50) and non-cancerous (n=50). The expression levels of MUC1 mRNA and miR-21marker in peripheral blood were higher in OSCC than in non-cancerous persons (p-value < 0.001) (Fig. 1). Afterward, we analyzed the expression of markers (MUC1 mRNA, miR-21), and the results showed that the MUC1 mRNA marker was positive in 35 of the 50 OSCC patients analyzed (70%). In non-cancerous it was 9 of the 50 persons (18%). The miR-21 was positive in 43 of the 50 OSCC patients (86%), and it was 14 of the 50 non-cancerous persons (28%).
Figure 1 : The expression levels of MUC1 mRNA and miR-21 in peripheral blood from OSCC patients and noncancerous persons
Correlations between the expression of MUC1 mRNA and miR-21
ΔCt (cycle threshold difference) values of MUC1 mRNA and miR-21 were compared respectively in cancerous and non-cancerous samples. Then, the difference in marker expression between the 2 groups was examined with the ΔΔCt method.
ΔΔCt = ΔCt sample – ΔCt calibrator
The computed ΔΔCt for MUC1 mRNA was -4.52 and for miR-21 was -4.2. Then, the expression relative of these markers was calculated as follows:
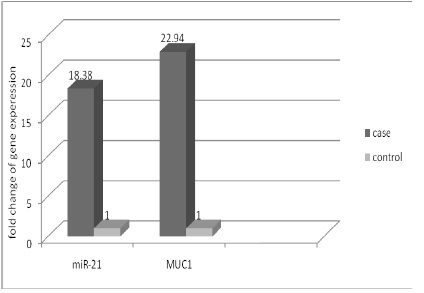
Relative expression = 2-ΔΔCt. The relative expression for the miR-21 marker in patients was 18.38times the non-cancerous persons, and for MUC1 mRNA marker were 22.94timesthe non-cancerous persons (Fig. 2).
Figure 2 :The difference in expression of miR-21 and MUC1 genes at the case and control
Elisa test results
Mean serum VEGF levels in patients with oral squamous cell carcinoma in peripheral blood were significantly higher than of those in the non-cancerous persons (p-value<0.001). The expression of VEGF marker was positive in 32 of the 50 OSCC patients analyzed (64%). This biomarker in healthy subjects reports not positive, indicating that the specificity is 100%.
Discussion
According to the WHO Globocan cancer statistics, oral cavity cancer occupies the 10th place in cancer morbidity for men on both the world statistics chart and the WHO European regional statistics. It has an age-standardized incidence rate of 3.8 on a world scale and 4.3 in Europe considering both sexes. The age-standardized mortality rates are 1.9 (world) and 1.7 (Europe), almost half of the rate of the incidences. Squamous cell carcinoma is responsible for approximately 90% of all malignant histopathological entities of the oral cavity(4). Oral squamous cell carcinoma (OSCC) is the eighth most common cancer worldwide, and its prevalence is high in Brazil, where 15.290 new cases of oral cancer were estimated to develop in 2014[25]. Oral squamous cell carcinoma is on the rise and cause of death of the human, therefore, earlier diagnosis is important to better prognosis and treatment, and tumor markers play a big role in the early detection of the disease. Our efforts in this study are to introduce methods for early diagnosis of this disease. In patients with OSCC expression of MUC1 mRNA, miR-21 and VEGF protein increase in serum and in other body fluids. In fact, this expression can be used as a diagnostic biomarker for OSCC, and we can use it as an agent useful for the detection of early prognosis and better treatment; so, biomarkers can be used with a simple blood test for cancer at the same stage diagnosed and treated early[26, 27, 28, 29, 30, 31). Earlier studies have shown that microRNAs (miR-21) play crucial roles in carcinogenesis, cellular processes, and disease mechanisms. Including cancer, and are considered as one of the most studied oncomiRNAs.They are also extensively involved in cancer pathogenesis of solid tumors and support their function as either dominant or recessive cancer genes [15, 32, 33]. Another study showed that a miR-21 expression was significantly associated with the degree of differentiation of tumor tissues, local invasion and lymph node metastasis[34]. One study investigated that the miR-21 expressed in the tumor stromal fibroblast and in some tumor vessels in OSCC patients[35]. One study reported miR-21 as a tumor oncogenic molecule and investigated that the expression of miR-21 in OSCC tissues was higher than that in noncancerous tissue [36]. One study was found that the expression of VEGF-A was high in OSCC patients with controls [19]. In another research, a correlation between VEGF serum and salivary levels has been revealed. These relationships are so important that researchers could be using this as a tumor marker[37]. We chose MUC1 mRNA as a marker to be tested, as MUC1 mRNA for diagnosis of lymph node micrometastases in non-small cell lung carcinoma. MUC1 mRNA was therefore evaluated by real-time RT-PCR in order to occult cancer cells[38]. The result of another study suggests that the determination of MUC1 expression may be a useful diagnostic marker for the OSCC[39]. MUC1 may be selected as a target for the effective treatment methods in OSCC(10). Based on this evidence, we can conclude that the biomarkers (miR-21, MUC1 mRNA and VEGF protein) were produced by tumor tissues. In addition, serum levels of these biomarkers in patients of OSCC compared to healthy subjects significantly increased.in Conclusion,According to our findings, as well as increased expression of these biomarkers in serum relates to tumor size and tumor staging. As a result, the increased expression of these biomarkers in the serum can be useful for early diagnosis of OSCC. However, further research is recommended to support further studies with a larger sample size.
Acknowledgments
The authors thank the Tehran University of Medical Sciences for this research.
References
[1]. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90.
[2]. Kawakita A, Yanamoto S, Yamada S-i, Naruse T, Takahashi H, Kawasaki G, et al. MicroRNA-21 promotes oral cancer invasion via the Wnt/β-catenin pathway by targeting DKK2. Pathology & Oncology Research. 2014;20(2):253-61.
[3]. Sessions DG, Lenox J, Spector GJ, Chao C, Chaudry OA. Analysis of treatment results for base of tongue cancer. Laryngoscope. 2003;113(7):1252-61.
[4]. Gombos K, Horvath R, Szele E, Juhasz K, Gocze K, Somlai K, et al. miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 2013;33(4):1511-7.
[5]. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4-5):309-16.
[6]. Cheng A, Schmidt BL. Management of the N0 neck in oral squamous cell carcinoma. Oral Maxillofac Surg Clin North Am. 2008;20(3):477-97.
[7]. Kowalski LP, Bagietto R, Lara JR, Santos RL, Silva JF, Jr., Magrin J. Prognostic significance of the distribution of neck node metastasis from oral carcinoma. Head Neck. 2000;22(3):207-14.
[8]. Capote A, Escorial V, Muñoz‐Guerra MF, Rodríguez‐Campo FJ, Gamallo C, Naval L. Elective neck dissection in early‐stage oral squamous cell carcinoma—does it influence recurrence and survival? Head & neck. 2007;29(1):3-11.
[9]. Mahfouz ME, Rodrigo JP, Takes RP, Elsheikh MN, Rinaldo A, Brakenhoff RH, et al. Current potential and limitations of molecular diagnostic methods in head and neck cancer. Eur Arch Otorhinolaryngol. 2010;267(6):851-60.
[10]. Kamikawa Y, Kanmura Y, Hamada T, Yamada N, Macha MA, Batra SK, et al. Combination of MUC1 and MUC4 expression predicts clinical outcome in patients with oral squamous cell carcinoma. Int J Clin Oncol. 2015;20(2):298-307.
[11]. Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, et al. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes & development. 2008;22(1):8-13.
[12]. Waldman SA, Terzic A. MicroRNA signatures as diagnostic and therapeutic targets. Clinical chemistry. 2008;54(6):943-4.
[13]. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-97.
[14]. Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6(2):127-38.
[15]. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257-61.
[16]. Ciafre S, Galardi S, Mangiola A, Ferracin M, Liu C-G, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and biophysical research communications. 2005;334(4):1351-8.
[17]. Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer research. 2005;65(14):6029-33.
[18]. Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr Rev. 1997;18(1):4-25.
[19]. Nayak S, Goel MM, Chandra S, Bhatia V, Mehrotra D, Kumar S, et al. VEGF-A immunohistochemical and mRNA expression in tissues and its serum levels in potentially malignant oral lesions and oral squamous cell carcinomas. Oral Oncol. 2012;48(3):233-9.
[20]. Li QL, Chen FJ, Zeng ZY, Yang AK, Wu QL, Zhang HZ, et al. [Expression and clinical significance of VEGF-C and Flt-4 in tongue squamous cell carcinoma]. Ai Zheng. 2006;25(2):235-40.
[21]. Martin SG, Orridge C, Mukherjee A, Morgan DA. Vascular endothelial growth factor expression predicts outcome after primary radiotherapy for head and neck squamous cell cancer. Clin Oncol (R Coll Radiol). 2007;19(1):71-6.
[22]. Siriwardena B, Kudo Y, Ogawa I, Udagama M, Tilakaratne W, Takata T. VEGF-C is associated with lymphatic status and invasion in oral cancer. Journal of clinical pathology. 2008;61(1):103-8.
[23]. Salven P, Heikkila P, Anttonen A, Kajanti M, Joensuu H. Vascular endothelial growth factor in squamous cell head and neck carcinoma: expression and prognostic significance. Mod Pathol. 1997;10(11):1128-33.
[24]. Artese L, Rubini C, Ferrero G, Fioroni M, Santinelli A, Piattelli A. Microvessel density (MVD) and vascular endothelial growth factor expression (VEGF) in human oral squamous cell carcinoma. Anticancer Res. 2001;21(1B):689-95.
[25]. Coutinho-Camillo CM, Lourenco SV, de Araujo Lima L, Kowalski LP, Soares FA. Expression of apoptosis-regulating miRNAs and target mRNAs in oral squamous cell carcinoma. Cancer Genet. 2015;208(7-8):382-9.
[26]. Karimi S, Mohamadnia A, Nadji SA, Yadegarazari R, Khosravi A, Bahrami N, et al. Expression of two basic mRNA biomarkers in peripheral blood of patients with non-small cell lung cancer detected by real-time rt-PCR, individually and simultaneously. Iranian biomedical journal. 2015;19(1):17.
[27]. Karimi S ,Bahrami N, Sharifi K, Daustany M, Baghbani- Arani F, Kazempour M, et al. . Investigating gene expression level of MUC1 and CEA in pleural fluid of NSCLC lung cancer patients with real-time RT-PCR method. MINERVA PNEUMOLOGICA. 2017 201718-24; 56(1):18-24.
[28]. Mohamadnia A, Karimi S, Yadegar Azari R, Naji SA, Khosravi A, Bahrami N, et al. Expression Of CK19 Gene In Patients With Lung Cancer And Its Comparison With Carcinoembryonic Antigen In Peripheral Blood. Journal of Payavard Salamat. 2016;9(5):459-68.
[29]. Bahrami N, Gholami M, Jamaati HR, Mohamadnia A, Dargahi H, Kazempour dizaji M,Khosravi A, Heshmatnia J, Vahabi P,Bahrami NA. Expression of two essential mRNA biomarker in the peripheral blood as possible biomarkers for diagnosis of non‑small cell lung carcinoma. MINERVA PNEUMOLOGICA. 2016;55(3):31-6.
[30]. Ghadimi K, Bahrami N, Fathi M, Farzanegan B, Naji T, Emami M, et al. D. Diagnostic value of LunX mRNA and CEA mRNA expression in pleural fluid of patients with non-small cell lung cancer. MINERVA PNEUMOLOGICA. 2017;56.
[31]. Moshref Behzad N ,Bahrami N, Farzanegan B, Fathi M, Zareh Karizi S, Mohamadnia A. Expression of CK19-mRNA and CEA -mRNA biomarkers in pleural fluid of patients with non-small cell lung cance. MINERVA PNEUMOLOGICA. 2017;56.
[32]. Zheng Y, Cui L, Sun W, Zhou H, Yuan X, Huo M, et al. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer biomarkers. 2012;10(2):71-7.
[33]. Selcuklu SD, Yakicier MC, Erson AE. An investigation of microRNAs mapping to breast cancer related genomic gain and loss regions. Cancer Genet Cytogenet. 2009;189(1):15-23.
[34]. Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncology reports. 2012;27(4):1019-26.
[35]. Hedback N, Jensen DH, Specht L, Fiehn AM, Therkildsen MH, Friis-Hansen L, et al. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One. 2014;9(4):e95193.
[36]. Wang W, Songlin P, Sun Y, Zhang B, Jinhui W. miR-21 inhibitor sensitizes human OSCC cells to cisplatin. Mol Biol Rep. 2012;39(5):5481-5.
[37]. Polz-Dacewicz M, Strycharz-Dudziak M, Dworzański J, Stec A, Kocot J. Salivary and serum IL-10, TNF-α, TGF-β, VEGF levels in oropharyngeal squamous cell carcinoma and correlation with HPV and EBV infections. Infectious Agents and Cancer. 2016;11(1):45.
[38]. Saintigny P, Coulon S, Kambouchner M, Ricci S, Martinot E, Danel C, et al. Real-time RT-PCR detection of CK19, CK7 and MUC1 mRNA for diagnosis of lymph node micrometastases in non small cell lung carcinoma. Int J Cancer. 2005;115(5):777-82.
[39]. Kumar MH, Sanjai K, Kumarswamy J, Keshavaiah R, Papaiah L, Divya S. Expression of MUC1 mucin in potentially malignant disorders, oral squamous cell carcinoma and normal oral mucosa: An immunohistochemical study. J Oral Maxillofac Pathol. 2016;20(2):214-8.
License
Copyright
© ,
Author Details