GSTM1 and GSTT1 Polymorphisms and Susceptibility to Acute Myeloid Leukemia: A Case-control Study of the Sudanese Population
Download
Abstract
Background: Glutathione S-transferase (GST) enzyme levels are associated with risk of many types of cancers, including hematological malignancies. In this study we here aimed to investigate the relationship between GSTM1 and GSTT1 polymorphisms and the risk of AML. Conflicts in the published results and the absence of similar in-depth studies in Sudan prompted us to perform the present case-control study to determine the frequency of GSTM1 and GSTT1 polymorphisms in AML patients and their possible association with AML in a Sudanese population.
Materials and Methods: A total of 40 patients with AML and 40 control subjects were enrolled in this study. Blood samples were collected from all patients in EDTA containing tubes. Genomic DNA was extracted from all blood samples using salting out method. Genotyping for detection of GSTM1, and GSTT1 polymorphisms was performed for both patients and controls using a multiplex PCR.
Results: We reported that there is an association between the GSTM1 null genotype and AML risk (OR= 2.7, 95% CI= 1.2-6.04; P.value = 0.012), the GSTT1 null genotype appeared also to have an influence in the development of AML (OR= 4.93, 95% CI= 1.6-15.07; P.value = 0.005).
Conclusion: These findings indicate that genetic variants of GSTM1 and GSTT1 genes may increase individual susceptibility to AML.
Introduction
Glutathione S-transferases (GSTs) are a supergene family of detoxifying enzymes which are found in virtually all life forms [1]. GSTs are phase II detoxifying enzymes that catalyze the conjugation of reduced glutathione with a wide variety of electrophilic substrates [2]. In addition to their function in xenobiotic detoxification, GSTs have peroxidase and isomerase activities that can inhibit the c-Jun N-terminal kinase (JNK) [3]. GSTs can also bind non-catalytically with a wide range of endogenous and exogenous ligands [3].
In humans, GST enzymes consist of many cytosolic, mitochondrial, and microsomal proteins, and the cytosolic family has eight distinct classes: alpha (A), kappa (K), mu (M), omega (O), pi (P), sigma (S), theta (T), and zeta (Z)[1][4].
Polymorphisms within genes that encode GSTs have been associated with susceptibility to nonmalignant[5]and malignant human diseases, [6] including acute myeloid leukemia (AML). [7]. Presumably, altered cancer risk because of polymorphic variation is mediated by differential ability to conjugate and detoxify both endogenously formed and exogenously derived electrophiles and their metabolites. Due to their multiple functions, polymorphic variants of GST enzymes may account for interindividual differences in outcome of chemotherapy. There are several reports on the role of GSTM1 and/or GSTT1 deletions as prognosticators in acute leukemia. [8][9].
To our best of knowledge this is the first study conducted in to determine the frequency of GSTM1 and GSTT1 polymorphisms in AML patients and their possible association with AML in a Sudanese population.
Materials and Methods
This is a descriptive case control study conducted in Khartoum state during the period from October 2014 to March 2015. Patients group was obtained at Flow Cytometry Laboratory Center at Khartoum, Sudan, where the patients were referred for immunophenotypic diagnosis.
Blood samples (3ml) were collected from 40 patients and 40 controls in ethylene diamine tetra acetic acid (EDTA) containers and genomic DNA was extracted by salting out method [10]. All patients and controls sampled in the study were consented verbally and the study was approved by the Human Ethics Committee of Al Neelan University.
Genotyping for detection of GSTM1, and GSTT1 polymorphisms was performed for both patients and controls using a multiplex PCR modified from Abdel-Rahman[11].
PCR amplification was carried out in 25 μl reactions containing three microliter (μl) of DNA , 1μl of each primer (Table 1), 4μl Matser mix (GoTaq® Green Master Mix, Promega, USA) and 12 μl of nuclease free water.
Thermal cycling conditions were as follows: an initial denaturation step at 95̊ C for 5 min, 35 cycles at 94̊ C for 1 min, 60̊ C for 1 min and 72̊ C for 1 min, and a final extension step at 72̊ C for 5 min (Table 1).
| PCR conditions | Primer Sequence | Primer direction |
| D: 95 °C 60 s A: 60 °C 60 s E: 72 °C 60 s 35X | 5'- GAACTCCCTGAAAAGCTAAAGC-3', | GSTM1 FP |
| 5'-GTTGGGCTCAAATATACGGTGG-3', ' | GSTM1RP | |
| 5'-TTCCTTACTGGTCCTCACATCTC-3', | GSTT1 FP | |
| 5'-TCACCGGATCATGGCCAGCA-3' | GSTT1RP | |
| 5'-CAACTTCATCCACGTTCACC-3', | ß-globin FP | |
| 5'-GAAGAGCCAAGGACAGGTAC-3 | ß-globin RP |
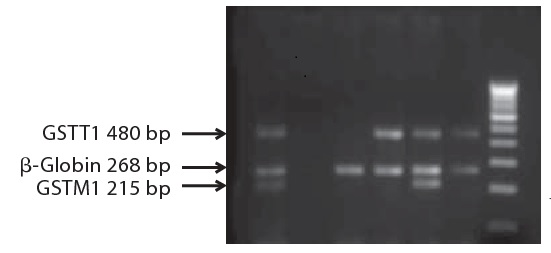
For analysis, 5 μl of each PCR product was fractionated on a 2% agarose gel with 0.05 μg/ml ethidium bromide. PCR products were directly visualized using UV fluorescence. GSTM1 & GSTT1 genotypes were determined by the presence and absence (null) of bands of 230 bp and 480 bp, respectively, with an internal control of 500 bp ( Figure 1) .
Figure 1: Agarose gel demonstrating multiplex PCR genotyping of GSTM1 and GSTT1 gene deletion (null genotype). The absence of a 480-bp band indicates the GSTT1 null genotype. The absence of a 215-bp band indicates the GSTM1 null genotype. ‐Globin was coamplified in all the samples. Lanes 1 and 5 represent the GSTM1 and GSTT1 positive genotype. Lanes 4 and 6: GSTM1 null genotype. Lane 2: the negative control, lane 3: the GSTM1 and GSTT1 null genotypes. M marker = 100-bp DNA ladder. .
Statistical analysis was carried out using Statistical Package for the Social Sciences version 21.0 software (SPSS) (Chicago, IL, USA). The odds ratio (OR) and its 95% confidence interval (CI) were used to illustrate the association between genetic variants and their risk for disease. The lowest accepted level of significance was 0.05 or less.
Results
The frequency of subjects carrying the GSTM1 null polymorphism was significantly higher in AML patients (60.7%) compared to controls (36 %) (OR= 2.7, 95% CI= 1.2-6.04, p.value= 0.012). For the GSTM1 normal genotype frequencies were 39.3 % in patients’ group and 64 % in controls’ group. (Table 2).
The frequencies of GSTT1 null genotype were higher in AML patients (60 %) than in control`s group (23.4%). However, for the GSTT1 normal genotype frequencies were 40 % in patients’ group and 76.7% in controls’ group. Statistically, significant difference was observed (OR= 4.93, 95% CI= 1.6-15.07; p.value= 0.005) (Table 2).
| Genotype | Group | Odd Ratio | p.value | 95% CI | |
| AML Patients | Controls | ||||
| GSTM1 Null | 60.7% | 36% | 2.7 | 0.012 | 1.2-6.04 |
| GSTM1 Present | 39.3% | 64% | |||
| GSTT1 Null | 60% | 23.4% | 4.93 | 0.005 | 1.6-15.07 |
| GSTT1 Present | 40% | 76.7% |
Discussion
Glutathione s-transferase (GST) polymorphisms (GSTM1, GSTP1 and GSTT1) have been reported to be risk factors for developing cancer and leukemia in many published studies; several studies reported the association between GSTT1 polymorphisms and risk of AML [12].
In this case-control study, we found that the GSTM1 and GSTT1 deletion polymorphisms were associated with risk of AML these findings are partially agree with study done in Egypt [13] which demonstrated that GSTM1 null or GSTT1 null genotypes may be considered independent risk factors for AML. This study indicated GSTs polymorphisms were promising candidate biomarkers for evaluating the AML risk. There are several previous studies investigated the association between GSTT1 polymorphisms and risk of AML [14][15][16][17], but the results are inconsistent. There is a case-control study conducted in the United States by Crump et al, and the results do not support the hypothesis that the GSTT1 gene deletion is related to the risk of AML [14]. Liu r also has reported that the variation in GSTT1 genotype is not associated with the susceptibility of risk of development of AML in a Chinese population [16].in Conclusion,Our finding indicates that genetic variants of GST s (GSTT1 and GSTM1 null genotypes) genes may increase individual susceptibility to AML.
Limitations
The study was limited by the small sample size even though it provides a preliminary data about the association of GSTT1 and GSTM1 null genotypes with susceptibility to AML. The study did not look further into the impact of the GSTs polymorphisms on the patient prognosis. The effect of the GST genotypes on the haematologial findings has to be studied further.
Declarations
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IKI carried out the molecular genetic studies and drafted the manuscript. NMA revised the manuscript. SOA participated in the patient enrolment. EAB performed the statistical analysis and revised the manuscript. RH revised the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
All original or analyzed data for this study is available on request from the corresponding author.
Ethics approval and consent to participate
The study was approved by the Human Ethics Committee of Al Neelan University. Informed consent was obtained from the participants.
Consent of publication
Not applicable.
Funding
This research did not receive any fund or financial support.
Acknowledgements
We are indebted to all the patients who participated in the study. We would also like to thank our technical colleagues (Molecular Biology Laboratory, Faculty of Medical Laboratory Sciences, Al Neelan University).
References
- The Glut athione S-Transferase Supergene Family: Regulation of GST and the Contribution of the lsoenzymes to Cancer Chemoprotection and Drug Resistance Part I Hayes John D., Pulford David J.. Critical Reviews in Biochemistry and Molecular Biology.1995;30(6). CrossRef
- GLUTATHIONE TRANSFERASES Hayes John D., Flanagan Jack U., Jowsey Ian R.. Annual Review of Pharmacology and Toxicology.2005;45(1). CrossRef
- GSTM1 and GSTT1 Polymorphisms and Susceptibility to Prostate Cancer: A Case-Control Study of the Algerian Population Benabdelkrim Maroua, Djeffal Omar, Berredjem Hajira. Asian Pacific Journal of Cancer Prevention.2018;19(10). CrossRef
- Glutathione-S-transferase family of enzymes Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Mutat Res.2001;482(1-2):21-26.
- Glutathione S-transferase M1 null genotype is associated with a decreased risk of myocardial infarction Wilson MH, Grant PJ, Hardie LJ, Wild CP. FASEB J.2000;14(5):791-796.
- Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, et al . Carcinogenesis.1997;18(7):1285-1289.
- Polymorphic variation within the glutathione S-transferase genes and risk of adult acute leukemia Rollinson S, Roddam P, Kane E, Roman E, Cartwright R, Jack A, et al . Carcinogenesis.2000;21(1):43-47.
- Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: A case-control study Stanulla M, Schrappe M, Brechlin AM, Zimmermann M, Welte K. Blood .2000;95(4):1222-1228.
- GlutathioneS-Transferase Polymorphisms and Outcome of Chemotherapy in Childhood Acute Myeloid Leukemia Davies Stella M., Robison Leslie L., Buckley Jonathan D., Tjoa Tom, Woods William G., Radloff Gretchen A., Ross Julie A., Perentesis John P.. Journal of Clinical Oncology.2001;19(5). CrossRef
- A simple salting out procedure for extracting DNA from human nucleated cells Miller S, Dykes D, Polesky H. Nucleic Acids Res.1988;16(3):1215.
- A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies Abdel-Rahman S, El-Zein R, Anwar W, Au W. Cancer Letters.1996;107(2):229-233.
- Risk Effects of GST Gene Polymorphisms in Patients with Acute Myeloid Leukemia: A Prospective Study Zhou L, Zhu Y, Zhang X, Li Y, Liu Z. Asian Pacific Journal of Cancer Prevention.2013;14(6):3861-3864.
- Glutathione S transferase (GSTP 1, GSTM 1, and GSTT 1) gene polymorphisms in Egyptian patients with acute myeloid leukemia Nasr AmlS, Sami RaniaM, Ibrahim NohaY, Darwish DaliaO. Indian Journal of Cancer.2015;52(4). CrossRef
- Glutathione S-transferase theta 1 gene deletion and risk of acute myeloid leukemia Crump C, Chen C, Appelbaum FR, et al . Cancer Epidemiol Biomarkers Prev.2000;9(5):457-460.
- Increased risk for acute myeloid leukaemia in individuals with glutathioneS-transferase mu (GSTM1) and theta 1 (GSTT1) gene defects Arruda VR, Lima CS, Grignoli CR, et al . Eur J Haematol.2001;66(6):383-388.
- Study on the relationship between polymorphisms of Cyp1A1, GSTM1, GSTT1 genes and the susceptibility to acute leukemia in the general population of Hunan province Liu QX, Chen HC, Liu XF, et al . Zhonghua Liu Xing Bing Xue Za Zhi.2005;26(12):975-979.
- Association ofGSTT1polymorphism with acute myeloid leukemia risk is dependent on smoking status Kim Hee Nam, Kim Nan Young, Yu Li, Tran Huong Thi Thanh, Kim Yeo-Kyeoung, Lee Il-Kwon, Shin Min-Ho, Park Kyeong-Soo, Choi Jin-Su, Kim Hyeoung-Joon. Leukemia & Lymphoma.2012;53(4). CrossRef
License
Copyright
© ,
Author Details