Hypoxia inducible factor-1 alpha (Hif-1 alpha) polymorphism in lymphoma and reactive lymphoid hyperplasia
Download
Abstract
Background: Lymphadenopathy is an abnormality in the size or the character of the lymph node. It may be a manifestation of infectious and malignant diseases. Reactive lymphoid hyperplasia is a benign form of lymphadenopathy. Cells develop numerous adaptive responses regulated by hypoxia inducible factor-1 alpha (Hif-1 alpha) against hypoxic stress.
Purpose: Hif-1alpha may play a role in the process of carcinogenesis in the early stage of cancer. We aimed to investigate the most common polymorphism of Hif-1α C1772T and G1790A gene polymorphisms in reactive lymphoid hyperplasia and lymphoma cases. Methods: Eighty-six paraffin-embedded blocs [51 (59,3%) reactive lymphoid hyperplasia; (40,7%) lymphoma] were examined. DNA was extracted from these samples and the polymerase chain reaction (PCR) was carried out. After DNA isolation, Hif-1α C1772T and G1790A polymorphisms were investigated with pyrosequencing.
Results: Cases were (29,1%) girls and 61 (70,9%) boys. The mean age was 91,47±57,96 and 142,46±41,66 for reactive lymphoid hyperplasia and lymphoma group, respectively. There was no Hif-1α C1772T gene polymorphism in both group, but Hif-1α G1790A gene polymorphism was recorded in 14 cases (reactive lymphoid hyperplasia 10, lymphoma 4). Although Hif-1α G1790A gene polymorphism was seen a little higher in reactive lymphoid hyperplasia cases than that of lymphoma, no meaningful relationship was found statistically between two groups (p>0,05).
Conclusion: Hif-1α C1772T and G1790A gene polymorphisms had been interrogated in cancer etiology and emphasized in some cancers. In our study, considering of a few of Hif-1α G1790A gene polymorphism in reactive lymphoid hyperplasia group, it should be investigated with large studies in terms of understanding of the behavior of Hif- 1α gene polymorphisms in children with lymphadenopathy.
Introduction
Lymphadenopathy is defined as an abnormality in the size or character of lymph nodes caused by the infiltration of inflammatory or malignant cells. Lymphadenopathy is usually due to local or systemic infection [1]. The most common diagnosis of lymph node enlargement is reactive lymphoid hyperplasia, which is a non-malignant and reversible enlargement of the lymph node due to antigenic stimuli such as infection or inflammation. But lymphadenopathy can also be a finding of malignant disease in children. The rate of malignant etiologies of lymphadenopathy is very low in childhood but increases with age [2].
Hypoxia is the most critical features of tumors. Hypoxic conditions induce some transcription factors for the maintenance of oxygen delivery to cancer cells. Hypoxia inducible factor-1 (Hif-1) is a transcription factor that plays a fundamental role in the physiological response to hypoxia. Hif-1 is composed of two subunits: Hif-1α and Hif-1β. Activation of Hif-1α is a significant driver of acute inflammation [3]. Hif-1α protein expression has been detected in most types of solid tumors and is associated with increased tumor growth, vascularization, and metastasis [4][5]. Hif-1α is overexpressed in colon, lung, and prostate cancers, but no expression was found in corresponding normal tissues, referring to the role of hif-1α in cancer [6]. The human Hif-1α gene, located at chromosome 14q21–24, codes for a 3919 bp cDNA and produces an 826 amino acid protein. Single nucleotide polymorphisms (SNPs) in coding regions can mediate amino acid changes and affect both the structure and biological activity of the translated protein. The most common investigation of Hif-1α polymorphisms is Hif-1α C1772T and G1790A gene polymorphisms. Under both hypoxic and normoxic conditions, these polymorphisms can cause increased transcriptional activity of Hif-1α [7].
It is well known that many genetic factors may have a role in childhood cancers. The most common cancers in children are leukemia and lymphoma. Many studies related to the role of Hif-1α in cancer showed that overexpression of Hif-1α has a prominent association with tumor development and metastasis. The association between Hif-1α C1772T and G1790A gene polymorphisms and various cancers had been studied. Tumor vasculature resulted in tumor progression caused by these polymorphisms was reported [8][9][10][11]. However effect of Hif-1α C1772T and G1790A gene polymorphisms did not address in lymph nodes in children. Therefore, we wanted to investigate Hif-1α gene polymorphisms that could be a biomarker for understanding molecular phenotypes of reactive lymphoid hyperplasia and lymphoma.
Materials and Methods
Ethical statement
This study was approved by the Ethics Committee at Yeditepe University, and written informed consents were obtained from each patient's family.
Study subjects
A total of 86 paraffin-embedded archival tissue blocks [51 (59,3%) reactive hyperplasia; 35 (40,7%) lymphoma] from the pathological files were obtained between years 2005 and 2010 in Medeniyet University Goztepe Education and Research Affiliated Hospital. All archive formalin-fixed paraffin-embedded tissue sections were prepared from lymph node biopsies. The lymphomas were classified using the WHO 2016 [12]. Demographic and clinicopathological data, including gender, age, histological lymphoma types, grades, and LDH levels were retrospectively collected by reviewing the patient charts. None of the patients had received preoperative radiotherapy or chemotherapy.
Sample collection and DNA isolation
Eighty-six FFPE tissue blocks from lymph node were cut into 5-10 μm section sand transferred to Yeditepe University School of Medicine Department of Medical Genetics. From these samples, DNA was extracted using the QIAamp DNA FFPE Spin Column Kit according to manufacturer protocol.
Polymerase chain reaction and pyrosequencing
The polymerase chain reaction (PCR) was carried out in a 25 μl reaction volume containing 2 μl of extracted DNA, 2 μl of forward primer (10 pmol/ml), 2 μl of reverse primer (10 pmol/ml), 12,5 μl of Master mixture, 1,5 μl Coral Load, 1,5 μl MgCl2, 2,5 μl Q solution and 3 μl of deionized water. The PCR primer sets were as follows Hif-1α, rs11549465 (C1772T) f:AGTTACGTTCCTTCGATCAGTTG; r:CTGCTGGAATACTGTAACTGTGCT and Hif 1α, rs11549467 (G1790A) f:CAGTTACGTTCCTTCGATCAGTTG; r:GACTTGCGCTTTCAGGGC (Table 1).
| Primer | Sequence (5’3’) | Length (bp) |
| Hif-1α, rs11549465 | f: AGTTACGTTCCTTCGATCAGTTG | 95 bp |
| r:CTGCTGGAATACTGTAACTGTGCT | ||
| Hif-1α, rs11549467 | f:CAGTTACGTTCCTTCGATCAGTTG | 67 bp |
| r:GACTTGCGCTTTCAGGGC |
Before pyrosequencing, all samples were loaded with 4 μl of amplified products and subjected to 40 min of electrophoresis under 120 V performed with 2% agarose gel containing 2 mg/ml ethidium bromide (EB) in a 0,5 x TAE buffer solution. The amplification bands were observed under ultraviolet (UV) light and photographed to check if the PCR was conducted successfully (Figure 1).
Figure 1: Agarose gel electrophrosesis of PCR products..

Detection of gene polymorphisms
A pyrosequencing method was designed to determine the genetic polymorphisms of Hif-1 alpha. Sequencing primer order of Hif-1α, rs11549465 (C1772T) and rs11549467 (G1790A) were GCGGAACTGCTTTCTAA and TGCGCTTTCAGGGCT, respectively. Pyrosequencing was carried out in a PyroMark Q24 (Qiagen, Germany), and the data were analyzed by using sequence analysis software.
Statistical Analyses
Statistical analyses were performed using SPSS 20.0 (SPSS Inc., Chicago, IL, USA). Descriptive analyses were presented using means and standard deviations (±1 SD) for continuous data and frequencies and percentages for categorical data. The variables investigated using Kolmogorov Smirnov test to determine whether or not they are normally distributed. Since the variables were normally distributed, Two independent samples t-test was used to compare the groups. Since the variables are not normally distributed, the Mann-Whitney U test was used to compare the groups. The Chi-Square and Fisher's exact test, where appropriate, was used to compare the proportions of the groups. A 5% type-I error level was used to infer a statistical significance. The level of significance was set at p<0,05.
Results
The total of 86 formalin-fixed paraffin-embedded tissue sections belonged to 51 (59,3%) reactive lymphoid hyperplasia and 35 (40,7%) lymphoma. The mean age was 112,22±57,49 months in 25 (29,1%) girls and 61 (70,9%) boys. Children diagnosed with reactive lymphoid hyperplasia were younger than that of lymphoma group. The correlation of two, according to the age, was statistically different (<0,001). The mean LDH level was 442,41±1133,42 U/L, and LDH was high in the lymphoma group.
Hif-1 alpha, rs11549467 (G1790A) gene polymorphism was found in 10 (71,4%) and 4 (28,6%) cases with reactive lymphoid hyperplasia and lymphoma group, respectively (Figure 2).
Figure 2: Hif-1α, rs11549467 (G1790A) gene polymorphism in reactive lymphoid hypeplasia and lymphoma group..
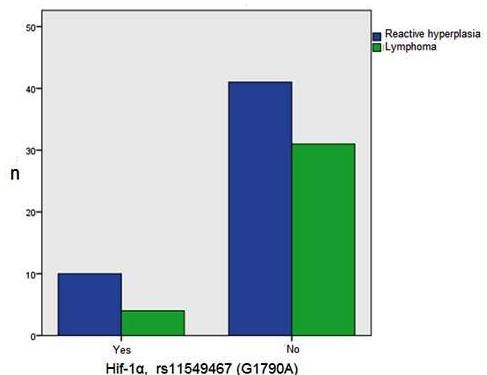
Lymphoma cases whose Hif-1 alpha, rs11549467 (G1790A) gene polymorphism were separated such as 3 Hodgkin's lymphoma (2 mixt cellularity type, 1 nodular sclerosing) and one case T cell non-Hodgkin lymphoma. Hif-1α, rs11549465 (C1772T) gene polymorphism was not detected in both groups. Although Hif-1α, rs11549467 (G1790A) gene polymorphism was seen mostly in reactive lymphoid hyperplasia, statistically difference was not found comparing of Hif-1α, rs11549467 (G1790A) gene polymorphism to both group (p=0.383) (Table 2).
| Reactive lymphoid hyperplasia group(percent) | Lymphoma group(percent) | p | |
| Number of cases | 51 (59,3) | 35 (40,7) | |
| Age (months) | 91,47±57,96 | 142,46±41,66 | <0,001 |
| Gender (F/M) | 12 (48,0)/39 (63,9) | 13 (52,0%)/22 (36,1) | 0,172 |
| LDH | 281,76±81,32 | 676,49±1762,608 | 0,194 |
| Hif-1 alpha, rs11549467 (G1790A) | 10 (71,4) | 4 (28,6) | 0,383 |
Of lymphoma tissues were 13 (37,14%) Non-Hodgkin and 22 (62,86%) Hodgkin lymphoma. The histological characteristics of Non-Hodgkin lymphoma were recorded as Burkitt lymphoma 5 (38,46%), T cell Lymphoblastic Lymphoma 7 (53,84%), Diffuse large B-cell lymphoma 1 (7,70%) and that of Hodgkin lymphoma were mixt cellular type 7 (31,82%), nodular sclerosing type 9 (40,90%), lymphocyte-rich type 6 (27,28%).
Lymphoma cases were presented such as 2 cases (5,7%) in stage I, 15 (42,9%) in stage II, 10 (28,6%) in stage III and 8 (22,9%) in stage IV. The duration of follow-up was 34,40±29,87 months in lymphoma cases. Hif-1α, rs11549467 (G1790A) gene polymorphism seen in both reactive lymphoid hyperplasia and lymphoma was pointed much more in boys than that of girls (Figure 3). But no association was found statistically (p=1000).
Figure 3: Hif-1α, rs11549467 (G1790A) gene polymorphism according to gender in reactive lymphoid hypeplasia (A) ve lymphoma (B) group..
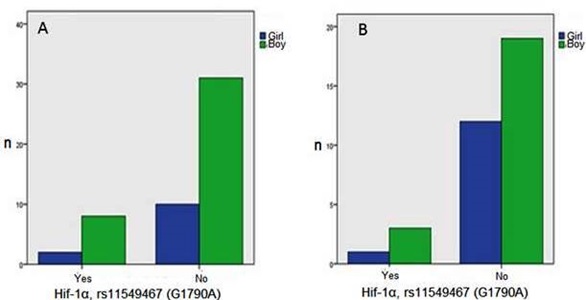
The comparison of Hif-1α, rs11549467 (G1790A) gene polymorphism and age (p=0,783), gender (p=1000) and LDH (p=0,861) levels were not significant statistically in reactive lymphoid hyperplasia and lymphoma cases (Table 3).
| Hif-1 α, rs11549467 (G1790A) | ||
| Reactive lymphoid hyperplasia(percent) | Lymphoma(percent) | |
| Age (month) | 81,20±56,69 | 148,75±29,15 |
| p=0,803 | p=0,783 | |
| Gender (F/M) | 2(20,0)/8(80) | 1(25)/3(75) |
| p=1,000 | p=1,000 | |
| LDH | 290,00±83,903 | 274,50±67,21 |
| p=0,722 | p=0,861 |
Hif-1α, rs11549467 (G1790A) gene polymorphism had no different significance statistically while comparing with histological types (p=0,726) and staging of lymphomas (p=0,383). Hif-1α G1790A gene polymorphism found only in 4 of 35 lymphoma cases were in remission. Furthermore, the meaningful difference between the duration of follow-up (p=0,900) and remission /relapse (p=1000) and Hif-1α, rs11549467 (G1790A) gene polymorphism was not found (Table 4).
| Lymphoma group | Hif-1 alpha, rs11549467(G1790A)(percent) | p |
| Histological types | ||
| Burkitt Lymphoma | 0(0,0) | |
| T cell ymphoblastic | 1(25,0) | |
| Lymphoma | 0(0,0) | 0,726 |
| Diffuse large B-cell lymphoma | 2(50,0) | |
| Mixed Cellular Type | 1(25,0) | |
| Nodular Sclerosing | 0(0,0) | |
| Lymphocyte Rich | ||
| Stage | 0,383 | |
| I | 0(0,0) | |
| II | 2(50,0) | |
| III | 0(0,0) | |
| IV | 2(50,0) | |
| Follow-up(months) | 39,00±28,40 | 0,900 |
| Remision/relaps | 4(100,0)/0(0,0) | 1,000 |
Discussion
Hif-1α C1772T and G1790A gene polymorphisms have been reported in many cancers including renal cell carcinoma, head and neck, and esophageal squamous cell carcinomas as well as colorectal and prostate cancers in previous studies [9]. These two polymorphisms have been considering in the diagnosis of some cancer types recently [13]. Meta-analysis studies showed that C1772T and G1790A polymorphisms of Hif-1α could be a potential marker for both cancer risk and cancer prognosis [14]. Immunohistochemical studies had shown that lymph node sections of non-Hodgkin's lymphoma had higher Hif-1α expression in adults [15]. In another study, Hif-1α expression was presented in lymphoma nodal samples with immunoblotting [16]. In our study, paraffin-embedded lymph nodes were examined for Hif-1α C1772T and G1790A polymorphisms with a pyrosequencing method. We conducted a comprehensive literature search to find a relationship between Hif-1α C1772T/G1790A polymorphisms and lymphoma in children. Until today these two polymorphisms not only lymphoma but also other cancers have never been studied in children. While we did not detect Hif-1α C1772T gene polymorphism, Hif-1α G1790A polymorphism was determined in the lymphoma group. Meta-analysis study performed by [17] examined the associations between HIF-1α C1772T polymorphism and the risk of cancer. They presented that many cancer types, including cervical cancer, colorectal cancer, breast cancer, oral cancer, renal cell carcinoma, and others had HIF-1α C1772T polymorphism except lymphoma. Further studies should be required to investigate the relationship between Hif-1α C1772T polymorphism and childhood tumors.
Intratumoral hypoxia and oncogenic mutations can cause to over-expressed of Hif-1α [6]. Previous studies performed the immunohistochemical expression of Hif-1α in various tumors. In one study, lymphoma tissues had weak Hif-1α expression compared to colon, lung, renal, and breast carcinomas. In the same study, interestingly immunohistochemical expression of Hif-1α was presented in germinal centers of reactive lymphoid tissues in 21 (84.0%) of the 25 of cases. The expression of Hif-1α more presented in germinal centers of reactive lymph nodes than that of parafollicular areas due to avascular germinal centers comparatively have hypoxic areas.
Consequently, germinal centers probably showed considerable Hif-1α expression [18]. Except that the expression of Hif-1α with immunohistochemically, both the C1772T and G1790A polymorphisms in the Hif-1α gene in reactive lymphoid tissues was not presented so far. We found only Hif-1α G1790A polymorphisms in 10 reactive lymphoid hyperplasia and 4 lymphoma cases. Even if G1790A gene polymorphisms of Hif-1a were not significantly associated with lymphoma and reactive lymphoid hyperplasia in our study, nodal samples with reactive lymphoid hyperplasia had more Hif-1α G1790A polymorphism than that of lymphoma tissues. Although the number of cases was very few, this result could be explained presumably by regarding much hypoxic areas of reactive lymphoid tissues than that of lymphomas. Besides, because of lymphoma patients whose Hif-1α G1790A polymorphism were few in our study, the comparison of lymphoma histology and stage with polymorphism should be strengthened by more cases.
Different meta-analysis studies are trying to explain the relationship between HIF-1α C1772T/G1790A polymorphisms and cancer risk [19][20]. In detail, meta-analysis indicated that both the polymorphisms 1772 C/T and 1790 G/A are significantly associated with overall cancer risk. Liu et al. presented in 8933 patients and 13,874 controls that the association between HIF-1α G1790A gene polymorphisms and cancer was significant [21]. Meta-analysis study suggests that Hif-1α gene G1790A polymorphism is a risk factor of cancer, especially for pancreatic cancer, lung cancer, renal cell carcinoma, and head and neck cancer [9]. In the light of these studies, Hif-1α could be a useful prognostic marker for early diagnosis of cancers. But the relationship between Hif-1α C1772T/G1790A gene polymorphisms and remission status of cancer did not display clearly. In one study, extramedullary infiltration was shown in acute myeloid leukemia with overexpression of Hif-1α [22]. We found Hif-1α G1790A gene polymorphism only in 4 of 35 lymphoma cases, and these cases were in remission. While Hif-1α gene G1790A polymorphism was considered as a risk factor for cancer, the explanation of remission status could be attributed to small samples in the present study. Therefore further studies are necessary to determine whether Hif-1α contributes to susceptibility to lymphoma growth or progression in children.
It is the investigation of whether Hif-1 alpha gene polymorphism is available or not in lymph nodes of children underwent biopsy due to suspicious lymphadenopathy. Our study has limitations. First, we included a few lymph node samples; our lymphoma cases might still be considered small. Second, we could not follow up with reactive lymphoid hyperplasia cases in a long time, and systemic changes in patient care cannot be entirely recorded. Third, the most significant limitation of our study is that we can not provide the other types of polymorphism of Hif-1α in lymph node samples.
In conclusion, although we had small samples, it should be thought that Hif-1α G1790A polymorphisms could play a critical role in children with lymphoma. Meanwhile, considering the finding of a relationship between reactive lymphoid hyperplasia and Hif-1α G1790A gene polymorphisms, children diagnosed with reactive lymphoid hyperplasia should be followed-up closely.
Acknowledgments
Funding
The present work was supported by a grant from TUBITAK (Project No: 2017-116S697).
Prior publication
No approved by any scientific body.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Ethical statement
This study was approved by the Ethics Committee at Yeditepe University, and written informed consents were obtained from each patient's family.
References
- Lymphadenopathy and malignancy Bazemore AW, Smucker DR. Am Fam Physician.2002;66(11):2103-2310.
- Peripheral lymphadenopathy: approach and diagnostic tools Mohseni S, Shojaiefard A, Khorgami Z, Alinejad S, Ghorbani A, Ghafouri A. Iran J Med Sci.2014;39(2):158-170.
- Activation of Hypoxia-Inducible Factor-1α in Type 2 Alveolar Epithelial Cell Is a Major Driver of Acute Inflammation Following Lung Contusion* Suresh Madathilparambil V., Ramakrishnan Sadeesh Kumar, Thomas Bivin, Machado-Aranda David, Bi Yu, Talarico Nicholas, Anderson Erik, Yatrik Shah M., Raghavendran Krishnan. Critical Care Medicine.2014;42(10). CrossRef
- Expression and correlation of hypoxia inducible factor HIF-1α and HIF-2α in cervical carcinoma Lixia J, Shaohua S, Qiaofa S, Feng L, Sijun L. J Pract Med.2012;28:1942-1943.
- Structural integration in hypoxia-inducible factors Wu Dalei, Potluri Nalini, Lu Jingping, Kim Youngchang, Rastinejad Fraydoon. Nature.2015;524(7565). CrossRef
- Expression of hypoxia-inducible factor 1: mechanisms and consequences Semenza Gregg L. Biochemical Pharmacology.2000;59(1). CrossRef
- HIF pathway mutations and erythrocytosis McMullin Mary Frances. Expert Review of Hematology.2010;3(1). CrossRef
- Correspondence Horrée N, Groot A J, van Hattem W A, Heintz A P M, Vooijs M, van Diest P J. Histopathology.2008;52(5). CrossRef
- Hypoxia-inducible factor-1α gene polymorphisms and cancer risk: a meta-analysis Zhao Tongfeng, Lv Jing, Zhao Jiangpei, Nzekebaloudou Marius. Journal of Experimental & Clinical Cancer Research.2009;28(1). CrossRef
- HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment Corzo Cesar A., Condamine Thomas, Lu Lily, Cotter Matthew J., Youn Je-In, Cheng Pingyan, Cho Hyun-Il, Celis Esteban, Quiceno David G., Padhya Tapan, McCaffrey Thomas V., McCaffrey Judith C., Gabrilovich Dmitry I.. The Journal of Experimental Medicine.2010;207(11). CrossRef
- Hypoxia inducible factor-1α gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma Hsiao Pei-Ching, Chen Mu-Kuan, Su Shih-Chi, Ueng Kwo-Chang, Chen Yi-Chen, Hsieh Yi-Hsien, Liu Yu-Fan, Tsai Hsiu-Ting, Yang Shun-Fa. Journal of Surgical Oncology.2010;102(2). CrossRef
- The 2016 revision of the World Health Organization classification of lymphoid neoplasms Swerdlow Steven H., Campo Elias, Pileri Stefano A., Harris Nancy Lee, Stein Harald, Siebert Reiner, Advani Ranjana, Ghielmini Michele, Salles Gilles A., Zelenetz Andrew D., Jaffe Elaine S.. Blood.2016;127(20). CrossRef
- Hypoxia-inducible factor-1 polymorphisms associated with enhanced transactivation capacity, implying clinical significance Tanimoto K.. Carcinogenesis.2003;24(11). CrossRef
- The association between HIF-1α polymorphism and cancer risk: a systematic review and meta-analysis Hu Xin, Fang Yuan, Zheng Jun, He Yazhou, Zan Xin, Lin Sen, Li Xi, Li Hao, You Chao. Tumor Biology.2013;35(2). CrossRef
- Level of PAX5 in differential diagnosis of non-Hodgkin′s lymphoma Mishra Rajnikant, Bharti Brij, Shukla Sachin, Tripathi Ratnakar, Mishra Suman, Kumar Mohan, Pandey Manoj. Indian Journal of Medical Research.2016;143(7). CrossRef
- Hypoxia inducible factor-alpha activation in lymphoma and relationship to the thioredoxin family Evens Andrew M., Schumacker Paul T., Helenowski Irene B., Singh Amareshwar T. K., Dokic Danijela, Keswani Anjeni, Kordeluk Elizabeth, Raji Adekunle, Winter Jane N., Jovanovic Borko D., Holmgren Arne, Nelson Beverly P., Gordon Leo I.. British Journal of Haematology.2008;141(5). CrossRef
- Hypoxia-Inducible Factor-1α C1772T Polymorphism and Cancer Risk: A Meta-Analysis Including 18,334 Subjects Ye Yanwei, Wang Min, Hu Shuang, Shi Yingqiang, Zhang Xiefu, Zhou Ye, Zhao Chunlin, Wang Guojun, Wen Jianguo, Zong Hong. Cancer Investigation.2014;32(4). CrossRef
- Expression of angiogenic factors and hypoxia inducible factors HIF 1, HIF 2 and CA IX in non-Hodgkin's lymphoma Stewart M, Talks K, Leek R, Turley H, Pezzella F, Harris A, Gatter K. Histopathology.2002;40(3). CrossRef
- Association between HIF-1α C1772T/G1790A polymorphisms and cancer susceptibility: an updated systematic review and meta-analysis based on 40 case-control studies Yan Qing, Chen Pin, Wang Songtao, Liu Ning, Zhao Peng, Gu Aihua. BMC Cancer.2014;14(1). CrossRef
- A meta-analysis of hypoxia inducible factor 1-alpha (HIF1A) gene polymorphisms: association with cancers Anam Md. T, Ishika Alokta, Hossain Md. B, Jesmin . Biomarker Research.2015;3(1). CrossRef
- 1790 G/A polymorphism, but not 1772 C/T polymorphism, is significantly associated with Cancers: An update study Liu Jie, Zhang Hong-xin. Gene.2013;523(1). CrossRef
- Expression of HIF-1α in primary acute myeloid leukemia cells and its relationship with prognosis Chen P, Jiang X, Huang HF, Yuan Q, Wu JY, Guo YF, et al . Zhongguo Shi Yan Xue Ye Xue Za Zhi.2015;23(1):19-23. CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2019
Author Details