Physicochemical Properties of analogs α-Aminophosphonates Drugs Determined via Molecular Dynamics Simulation
Download
Abstract
Objective: Cyclin-dependent kinases (CDKs, play important roles in cell cycle regulation. Since deregulation of cyclins and/or alteration or absence of inhibitors has been associated with many cancers, there is strong interest in CDKs inhibitors that could play an necessary role in the discovery of a new family of antitumor agents.
Material and Methods: Molecular modeling is used to design new materials, which the correct prediction of physical properties of realistic systems is required. Gromacs is an tool to perform molecular dynamics simulations and energy minimization of bimolecular systems which commonly consists of several tens to thousands of amino acid residues. From the simulations the first 100 ps were regarded as equilibration, leaving 900 ps for analysis purposes.
Results: Thermodynamic properties density, potential energy, temperature, and pressure are given. In this results tetramethyl ((1,4-phenylenebis (azanediyl)) bis ((4-chlorophenyl) methylene)) bis (phosphonate) was found to be the better selective known inhibitor for cyclin-dependent kinase2 because it shown lowest energy.
Conclusion: The phosphoric acid moiety is considered to bind to the affected protein more actively than the corresponding carboxylic acid because of its di anionic character.
Introduction
Understanding the function of biomolecules using the available structural data is one of the grand objections of the current century. The problem is too complex for analytical approaches; thus, progress in the field has been made principally via computer simulations. To rapid developments in computer technology, the growth in the field, in particular in molecular dynamics (MD) simulations, has been phenomenal [1]. From the early 1980s, three MD simulation packages with their associated force fields have prominent themselves, namely, AMBER, CHARMM and GROMACS. At present most of the simulation work on biomolecules is carried out using one of these packages.The force fields frequently used in molecular dynamics simulations of proteins are optimized under bulk conditions [2].
Cyclin-dependent kinases (CDKs), specially CDK1, CDK2, CDK4, and CDK6, play important roles in cell cycle regulation. CDK misregulation is indicated in a number of disease states, and in particular cancer. To advance such studies, small molecule cyclin-dependent kinase inhibitors would be worthy biochemical tools [3]. The cyclin-dependent kinases (CDKs) are a conserved family of proline-directed serine/threonine kinases that play a key role in regulation of cell cycle in eukaryotic cells [4].
The activity of CDKs is regulated most acutly through interaction with cyclins [5]. In addition, CDK also show a role in apoptosis (CDK2) and in the control of transcription (CDKs7, 8, 9) [6]. Protein kinases, through phosphorylation reactions, are involved in the regulation of various cellular activities since they belong to the cellular signaling molecules and therefore regulate all the processes that are basic for growth, development and homeostasis of eukaryotic cells [7]. Since deregulation of cyclins and/or alteration or absence of inhibitors has been associated with many cancers, there is strong interest in CDKs inhibitors that could play an necessary role in the discovery of a new family of antitumor agents [8].
α-Aminophosphonates are considered to be the structural analogs of the corresponding esters of α-amino acids and have been reported to expend several pharmacological activities such as their potential usage as anticancer drugs, enzymes inhibitors, haptens of catalytic antibodies, pharmacologic agents, antifungals, insecticides, plant growth regulators, and anti-HIV agents [9].
Materials and Methods
Molecular dynamics becomes the most effective way to express the details of the flow and to study many fundamental Nano fluid problems, which can be acutely difficult to investigate by other means [10].
Molecular modeling is used to design new materials, which the correct prediction of physical properties of realistic systems is required (Figure1).
Figure 1: 1GIH 126-132 residues, with colors that vary according to the residues type. Figure produced by VMD- software..
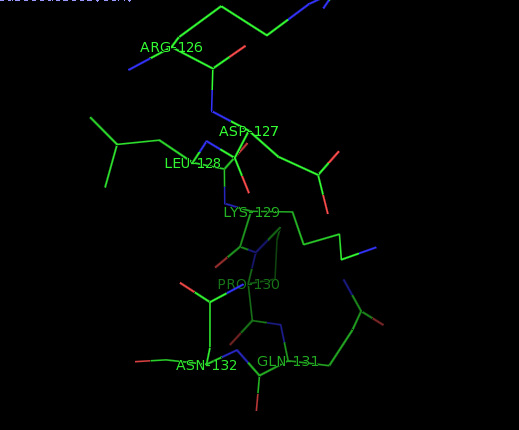
Gromacs is an tool to perform molecular dynamics simulations and energy minimization of bimolecular systems which commonly consists of several tens to thousands of amino acid residues [11].
In the present study, the gromacs software was employed for docking synthetic compounds into a protein, cyclin-dependent kinase 2 (PDB ID: 1GIH). GROMACS is an engine to perform molecular dynamics simulations and energy minimization. These are two of the many techniques that belong to the realm of computational chemistry and molecular modeling.
Different simulations were performed using Groningen machine for chemical simulations (GROMACS) 4.5 package.
From the simulations the first 100 ps were regarded as equilibration, leaving 900 ps for analysis purposes. Thermodynamic properties density, potential energy, temperature, and pressure are given.
Diffusion coefficient (D), represents the mean square displacement (MSD) and was calculated by GROMACS program g_msd.
Results
By reason of highly commercial and pharmaceutical importance and also to understand the biochemical and molecular basis of the biosynthesis of the natural compound is an interesting exercise [12].
These have considerable success in applying such basic screening approaches in the development of more potent and selective inhibitors against a range of different targets [13].
Hydrogen bonding is essential for the conformation and function of proteins. Proteins respond to change in pressure and temperature by diversity the number of hydrogen bonds with water [14].
The potential energy can be calculated as bonded (intra-molecular) and non-bonded (intermolecular) interactions. There are two potential functions to be concerned about between non-bonded atoms: Columbic interactions between electrostatic charges and Lennard- Jones interaction energy that reflects the van der Waals interaction between atoms. There are four types of interaction of bonded atoms: stretching along the bond and represents the energy required to stretch or compress a covalent bond, bending between bonds, planar distortion, and torsion [15].
The next simulations were done on the charged and neutral drugs (Table 1) in different dosages, in the presence of water molecules, and in the immediate vicinity of the interface of the CDK2.
| Number of Conformation | Inhibitor | Ki b | Ref RMS |
| (1a) | tetramethyl((1,4-phenylenebis(azanediyl))bis((4-(diethylamino)phenyl)methylene))bis(phosphonate) | 521.42 | 42.23 |
| (1d) | tetramethyl((1,4-phenylenebis(azanediyl))bis((4-chlorophenyl)methylene))bis(phosphonate) | 166.56 | 41.91 |
a) Kcal/mol
The GROMOS force field, modified by Berger, and introduced as an appropriate force field, was also used in the calculations (Figure 2).
Figure 2: The Density, Energy Plot Obtained at 300 K Temperature, the Root Mean Square Deviation, the rdf During 1000 Pico by Gromacs Tool During Simulation Corresponding to a Stabilized Structure of CDK2 (PDB code 1GIH),Second Molecular Dynamics; Simulation of the model at 300 K temperature by Gromacs tool. The Root mean square deviation plot obtained from Gromacs tool during molecular dynamics simulation for 1000 Pico second. The rdf plot obtained from Gromacs tool during molecular dynamics simulation for 1000 Pico second..
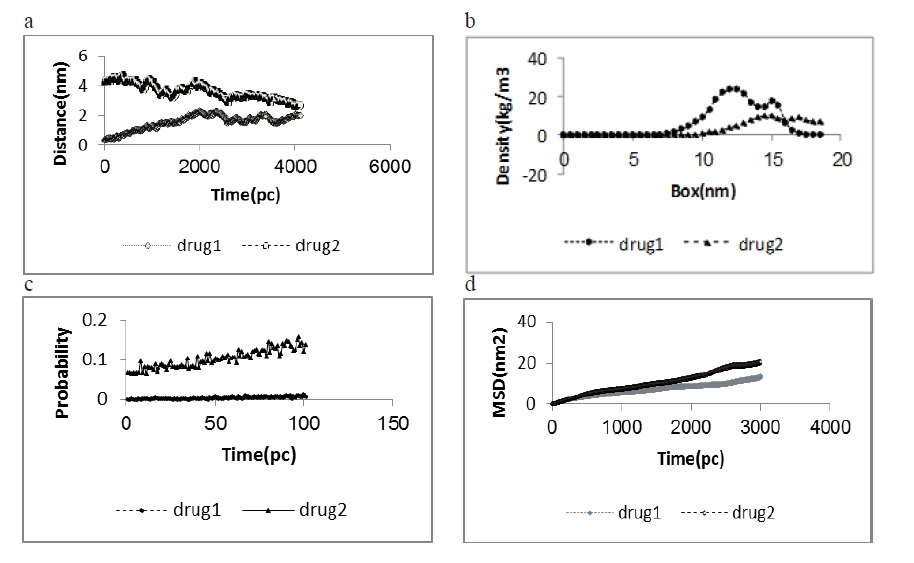
This analysis for determining the hydrogen bonding is based on the cutoffs for the acceptor-donor-hydrogen angle and the hydrogen-acceptor distance. Oxygen always acts as an acceptor, and the functional groups, such as NH and OH, act as donors.
In this results tetramethyl ((1,4-phenylenebis (azanediyl)) bis ((4-chlorophenyl) methylene)) bis (phosphonate) was found to be the better selective known inhibitor for cyclin-dependent kinase2 because it shown lowest energy.
Assuming that the system comprises of N atoms with mass {mi | i = 1, . . . ,N} at positions
{_ri | i = 1, . . . ,N}, the Hamiltonian of the total system is written as [16] (Figure 3):
Figure 3.
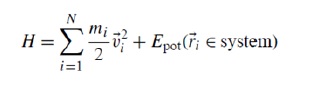
A great deal of work has gone into optimization of the parameters preferred in these force fields over the last 2 decades. Despite all this effort, one aspect of the force fields just mentioned has persisted basically untouched, namely, neglect of the polarization interaction.
Unlike the Coulomb and Lennard-Jones interactions, which are two-body and can be simply attained, the polarization interaction has a many-body character and requires a computationally expensive iteration process for implementation. The computer power in the early 1980s was simply not enough for explicit involvement of the polarization interaction. Since they cannot be declined absolutely, effects of polarization were included in a mean field sense by increasing the permanent dipole moments of molecules.
Discussion
Computer simulation has arrived as a particularly valuable tool for characterizing biomolecule conjugates due to the difficulty in obtaining actual experimental data for such systems [17].
Computational chemistry is just a name to make the use of computational techniques in chemistry, ranging from quantum mechanics of molecules to dynamics of large complex molecular aggregates. Molecular modeling indicates the general process of depicting complex chemical systems in terms of a realistic atomic model, with the goal being to understand and predict macroscopic properties based on detailed knowledge on an atomic scale.
Analysis of the number of hydrogen bonds between inhibitors and CDK2 shows that the CDK2–tetramethyl ((1,4-phenylenebis (azanediyl)) bis ((4-chlorophenyl) methylene)) bis (phosphonate) complex has properties, which indicates that it has higher affinity for CDK2 than other. Further inhibition experiments can confirm this prediction (Figure 2).
In view of the hypothesis dependence of the co-operatively on the conformational states of enzyme, and because of the drastic effects water can sometimes have on protein the Cyclin-dependent kinase reaction in H2O [18].
The phosphoric acid moiety is considered to bind to the affected protein more actively than the corresponding carboxylic acid because of its di anionic character.
Acknowledgments
We gratefully acknowledge the financial supports received for this research work from Niroo Research Institute. Furthermore, we would like to thank the Dr. Abbas Yousefpour for their generous support.
References
- Partitioning of Nonsteroidal Antiinflammatory Drugs in Lipid Membranes: A Molecular Dynamics Simulation Study Boggara Mohan Babu, Krishnamoorti Ramanan. Biophysical Journal.2010;98(4). CrossRef
- Interaction of Salicylate and a Terpenoid Plant Extract with Model Membranes: Reconciling Experiments and Simulations Khandelia Himanshu, Witzke Sarah, Mouritsen Ole G.. Biophysical Journal.2010;99(12). CrossRef
- Cyclin-Dependent Kinase Inhibition by New C-2 Alkynylated Purine Derivatives and Molecular Structure of a CDK2−Inhibitor Complex Legraverend Michel, Tunnah Paul, Noble Martin, Ducrot Pierre, Ludwig Odile, Grierson David S., Leost Maryse, Meijer Laurent, Endicott Jane. Journal of Medicinal Chemistry.2000;43(7). CrossRef
- Cyclin-Dependent Kinase Inhibitors as Marketed Anticancer Drugs: Where Are We Now? A Short Survey Mariaule Gaëlle, Belmont Philippe. Molecules.2014;19(9). CrossRef
- Virtual screening studies reveal linarin as a potential natural inhibitor targeting CDK4 in retinoblastoma Vetrivel Umashankar, Sivashanmugam Muthukumaran, Raghunath Chandana. Journal of Pharmacology and Pharmacotherapeutics.2013;4(4). CrossRef
- Effects of the nonsteroidal anti-inflammatory drug naproxen on human erythrocytes and on cell membrane molecular models Manrique-Moreno Marcela, Suwalsky Mario, Villena Fernando, Garidel Patrick. Biophysical Chemistry.2010;147(1-2). CrossRef
- Design and one-pot synthesis of α-aminophosphonates and bis(α-aminophosphonates) by iron(III) chloride and cytotoxic activity Rezaei Zahra, Firouzabadi Habib, Iranpoor Nasser, Ghaderi Abbas, Jafari Mohammad Reza, Jafari Abbas Ali, Zare Hamid Reza. European Journal of Medicinal Chemistry.2009;44(11). CrossRef
- Structural bioinformatics study of cyclin-dependent kinases complexed with inhibitors Canduri F., Silveira N. J. F. da, Camera Jr J. C., Azevedo Jr W. F. de. Eclética Química.2003;28(1). CrossRef
- Synthesis, characterization, and application of [1-methylpyrrolidin-2-one-SO3H]Cl as an efficient catalyst for the preparation of α-aminophosphonate and docking simulation of ligand bond complexes of cyclin-dependent kinase 2 Mirzaei Mahdi, Eshghi Hossein, Hasanpour Maede, Sabbaghzadeh Reihaneh. Phosphorus, Sulfur, and Silicon and the Related Elements.2016;191(10). CrossRef
- Molecular Dynamics Simulation of Water in Single WallCarbon Nanotube Farhadian1 N, Shariaty-Niassar M. International Journal of Nanoscience and Nanotechnology..
- Free energy calculations of protein-water complexes with Gromacs Shahzad M.A . bioRxiv preprint first posted online Apr. 21, 2018..
- Homology Modelling of Lycopene Cleavage Oxygenase: The Key Enzyme of Bixin Production Raghunath Satpathy, Rajesh Ku.Guru , Rashmiranjan Behera , Aparajita Priyadarshini. Journal of Computer Science & Systems Biology, Journal of Computer Science & Systems Biology..
- In Silico Docking, Molecular Dynamics and Binding Energy Insights into the Bolinaquinone-Clathrin Terminal Domain Binding Site Abdel-Hamid Mohammed, McCluskey Adam. Molecules.2014;19(5). CrossRef
- Physicochemical properties of blue fluorescent protein determined via molecular dynamics simulation Krasnenko Veera, Tkaczyk Alan H., Tkaczyk Eric R., Mauring Koit. Biopolymers.2008;89(12). CrossRef
- Computational Investigations of Biomolecular Systems and Comparison with Experiments in Various Environmental Conditions, Nove Hrady, Czech Republic, 2011 Morteza Khabiri. .
- Hybrid quantum mechanical/molecular dynamics simulation on parallel computers: density functional theory on real-space multigrids Ogata Shuji, Shimojo Fuyuki, Kalia Rajiv K., Nakano Aiichiro, Vashishta Priya. Computer Physics Communications.2002;149(1). CrossRef
- Model-free simulation approach to molecular diffusion tensors Chevrot Guillaume, Hinsen Konrad, Kneller Gerald R.. The Journal of Chemical Physics.2013;139(15). CrossRef
- Solvent isotope effects on the glucokinase reaction. Negative co-operativity and a large inverse isotope effect in 2H2O POLLARD-KNIGHT Denise, CORNISH-BOWDEN Athel. European Journal of Biochemistry.1984;141(1). CrossRef
License
Copyright
© ,
Author Details