Examining the Expression of miR-205 and CEA mRNA in Peripheral Blood of Patients with OSCC(Oral Squamous Cell Carcinomas) and Comparing them with Healthy People
Download
Abstract
Introduction: Cancer is one of the most important causes of mortality in the world. So, in this study the changes of expressing miR-205 and CEA in oral cancer in peripheral blood were examined for early detection and better treatment.
Methods: In this study, we selected the number of 30 patient people and 30 healthy people. We measured their blood miR-205 and CEA using Real-Time PCR technique and evaluated the relationship between the expression of these biomarkers with tumor staging and cancer progression.
Findings: there is no a significant difference in mean age by comparing these two groups using t-test. The CEA mRNA biomarker was positive in 24 out of 30 people of the patient people group and was positive in 4 out of 30 people of the healthy people group. Statistical comparison represented a statistically significant difference between the two groups (P-value <0.001). The miR-205 biomarker was positive in 9 out of 30 people of the patient people group and was positive in 22 out of 30 people of the healthy people group. Statistical comparison represented a statistically significant difference the two groups (P-value <0.001).
Conclusion: In general, the research result can be considered as a screening test for early detection of the disease in the early stages. It is recommended to conduct more extensive studies with larger sample sizes to further proof of the research results.
Introduction
Oral squamous cell carcinoma (OSCC) involves 95% of all oral cancers. Also, OSCC is the sixth most common malignancy in the world and responsible for 3% of all cancers, and is one of the ten most common causes of mortality in the world [1].
Generally, Oral cancer is spread through the lymphatic system (metastasis) and the cervical lymph nodes are often the closest place of their involvement [2].
Head and neck squamous cell carcinoma (HNSCC) involves a heterogeneous group of malignancies that include the oral cavity, nasal cavity, paranasal sinuses, pharynx, larynx, and salivary glands. In spite of recent advances in diagnosis and treatment methods, less than 50% of patients with OSCC survive for 5 years [3-4].
Most oral cancers are diagnosed in advanced stages. Indeed, these lesions are discovered when they cause to emerge the clinical symptoms manifestations due to high progression, this leads to poor oral cancer prognosis in most parts of the world [5]. In general, this cancer accounts for 5% of all cancers in men and 2% in women. It has been recorded that some carcinogens increasing the riske of this cancer in old ages, including smoking, alcohol and tobacco, increase the DNA damage as well as viruses and other microbial agents and their effects on the oral mucosa [6].
Biomarkers are biomolecules that are found in blood or other body fluids or tissues, representing a sign of a normal or abnormal process and/or a specific situation or disease [7-8]. Examining several biomarkers together can provide with medical staff more accurate and reliable results to diagnose the cancers [9].
MicroRNAs are a large subgroup of non-coding RNAs of 18-25 nucleotides. Interacting microRNAs with target genes characterizes their role in growth, planed death, cellular differentiation and proliferation, and confirms the direct function of microRNAs in cancer [10]. These molecules control the gene expression after transcription by inhibiting translation of mRNA or inducing its degradation [11-13]. Increasing or decreasing changes in the expression of some microRNAs, which lead to the cancer process, influence cell growth by interfering with cell cycle regulators [14-15]. The amount of miR-205 is decreased in breast tumor tissues, which is consistent with previous reports. Above all, Inappropriate expression of miR-205 increases significantly apoptosis and independent growth of breast cancer cells [16]. Carcinoembryonic antigen plays a role in cell adhesion and is usually synthesized during embryonic development and is stopped shortly before birth. Increasing the serum levels of CEA provide prognostic information about the course of the disease [17]. CEA plays an important role not only in the diagnosis but in helping to determine the stage of cancer, follow-up treatment and determine the cancer prognosis [18].
Materials and Methods
The number of 30 patients referred to Cancer Institute of Tehran University of Medical Sciences were selected based on physical examinations and diagnosis of OSCC by a medical specialist before carrying out any treatment. The number of 30 healthy people participated in the study as control group after voluntary examination and filling out a consent form. The samples of patient and healthy people included peripheral blood. People were considered in the same groups in terms of afe factor with minimum age of 22 years and maximum age of 77 years.
Then, 2 ml of peripheral blood was taken through a standard blood syringe in glass test tubes. And it immediately went into RNA extraction.RNA extraction was carried out using RNA Blood Mini Kit (qiagen Cat no.52304).
Viva 2-steps RT-PCR Kit (Cat no.RTPL12) was used to create the cDNA. Real-Time RT-PCR was used to examine the CEA gene using CinnaGreen qPCR Mix, 2X (Cat No.MM2041) (Table 1). The 18srRNA reference gene was used for the CEA mRNA biomarker. Primers F and R for CEA were GTGCCCCCTAGCAGTACCG and GACGTGCCCCTACAAGTTGG, respectively.
| Cycles | Duration of cycles | Temperature |
| 1 | 15 min | 95ºC |
| 35-40 | 15-30 seconds 60 seconds | 95ºC 55-60 ºC |
| 1 | Melting Analysis | 55-95 ºC |
Also, ZIST ROYESH kit was used for making cDNA and performing real-time RT-PCR and was carried out with Rotor-Gene –QIAGEN instrument. The reaction temperatures and times were adjusted according to the kit instruction. After completing of each reaction, the results were interpreted based on the amplification and melting peak curves.
Results
As abovementioned, the studied population was consisted of 30 healthy people and 30 patient with OSCC. These two groups were matched in terms of age variables. The groups were compared by means of t-test in terms of mean age and there was not a significant difference in terms of mean age; so it can be concluded that age factor is not a problem in the studied groups (Table 2).
| Age (years) | |||
| Main group | The age range | mean | Standard deviation (SD) |
| Patient (30 people) | 70-26 | 25/46 | 22/1 |
| Healthy (30 people) | 70-25 | 84/47 | 12/12 |
| SD=standard deviation | P-value=0.442 |
Analyzing the Expression of the Studied Biomarkers
The CEA mRNA biomarker was positive in 24 out of 30 patients, representing a sensitivity of 80%. The rate of this biomarker was 4 out of 30 people in the healthy group (Figure 1). Statistical comparison was performed on the rate of being positive of this biomarker in patient people and healthy people groups by Two-sample binomial test, representing a statistically significant difference between these two groups (P-value <0.001).
Figure 1: The Level of Experssion of CEA-mRNA and miR-205 in Peripheral Blood from Patient with OSCC and Healthy People..
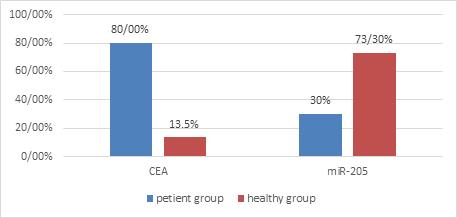
The miR-205 biomarker was positive in 9 out of 30 people in patient people group. The rate of this biomarker was 22 out of 30 people in the healthy people group (Figure 1). Statistical comparison was performed on the rate of being positive of this biomarker in patient people and healthy people groups by Two-sample binomial test, representing a statistically significant difference between these two groups (P-value <0.001).
Calculating the difference of expression level of biomarkers in two research groups
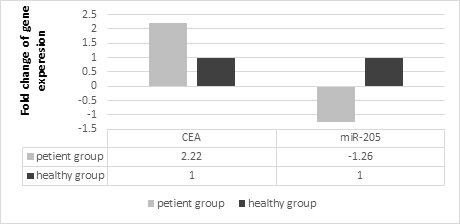
We used ΔΔ Ct method for this purpose. The ΔΔ Ct values for miR-205 and CEAmRNA biomarkers were 0.34 and 1.15, respectively.
Then, we used the 2-ΔΔCt formula. Therefore, if obtained ΔΔ Ct for the miR-205 biomarker put in this formula, the number of primary copies of this biomarker in healthy people is 1.26 times that of patients with cancer. And if we put the obtained ΔΔ Ct for the CEA mRNA biomarke in this formula, the number of primary copies of this biomarker in patients is 2.22 times that of healthy people (Figure 2).
Figure 2: The Difference between miR-205 and CEA Genes in the Case and the Control.
Discussion
Oral cancer accounts for about 4% of all body malignancies, and squamous cell carcinoma involves more than 90% of oral cavity cancers. This cancer has a high degree of local invasion and metastasis and enhances the mortality of patients [19].
OSCC accounts for 90% of all oral cancers. Also, it is the sixth most common malignancy in the world and responsible for 3% of all cancers, and is one of the ten most common causes of mortality in the world. In spite of advances in the field of surgery and radiotherapy to treat this malignancy, 5-year survival rate for this disease has not increased significantly. If the malignancy is detected in early stages, the survival rate is between 60-80% [1].
Hence, identifying and applying the molecular markers in the early detection of OSCC can help prevent and treat this cancer. Today, cancer cells can be detected in specific tissues by evaluating the expression of mRNA biomarkers. These biomarkers can be obtained from cancerous cells in the peripheral blood [20].
Almost during three to four decades, changes in the protein making of tumor suppressor genes or oncogenes have been considered as the major factor of tumor growth. However, the recent discovery of thousands of genes, which transcribe non-coding RNAs (including miRNAs), indicate that cancer biology is even more complex than initial expectations. Several levels of regulator molecules (e.g., mRNA, miRNA and protein) are involved in the development and maintenance of cancer phenotypes [21-22].
The usage of quantitative Real-time PCR is increasing in many diagnostic and molecular laboratories and is a good alternative to conventional PCR [23-24].
Vizcarra used Radioimmunoassay (RIA) in his studies and worked on TPA, CA15.3 and CEA. Also, he observed increasing these markers in patients. According to the Radioimmunoassay (RIA) method is less sensitive than Real-Time PCR, so Real-time PCR-based studies can characterize CEA as an appropriate molecular marker for early detection of breast cancer [25].
In a similar study, Adams et al. compared the tissue and serum VEGF. In this study, there was a significant relationship between the VEGF levels of all cancer groups (localized and metastatic) [26]. In the conducted study, there was a significant relationship between cancer and healthy groups.
In a study, Zhu G et al. examined the expression of CK19-mRNA and CEA-mRNA in the peripheral blood of NSLC patients and it was found that the positive expression rates for CK19-mRNA and CEA-mRNA were 57%, 40%, respectively, and was 43% for both. They have concluded that CK19-mRNA and CEA-mRAN are appropriate markers for dignosis of micro-metastasis [27]. In the present study, there was a significant difference between the CEA mRNA and miR-205 markers in the healthy and cancer groups.
Iorio and colleagues used a miRNA microarray in order to evaluate the miRNA expression from the specifications of 10 normal and 76 neoplastic breast tissues. They found that miR-10b, miR125b and miR-145 were reduced in breast cancer while miR-21 and miR-155 were increased in. Based on the microarray expression experiments, they found that miR-155 was highly expressed in BC while miR-145, miR-335, miR-10b, miR-125a and miR-205 were reduced [28].
In a study by Liu Jingjing et al. using Real-Time PCR (RT-PCR), two biomarkers miR-205 and miR-155 were investigated in the serum of 30 participants with breast cancer and 10 healthy people. The results indicated that miR-205 was reduced (downregulate) in serum of patients with breast cancer (BC), while miR-155 was increased (upregulate). This results are consistent with the results of the current study on miR-205 [29].
Acknowledgements
The authors thank the professors of Tehran University of Medical Sciences, Shahid Beheshti University of Medical Sciences and Islamic Azad University, Faculty of Biological Science.
References
- Cancer genes and the pathways they control Vogelstein Bert, Kinzler Kenneth W. Nature Medicine.2004;10(8). CrossRef
- Cancer Prevention Overview (PDQ®). 2017 Screening P, Board PE. .
- Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012 Ferlay Jacques, Soerjomataram Isabelle, Dikshit Rajesh, Eser Sultan, Mathers Colin, Rebelo Marise, Parkin Donald Maxwell, Forman David, Bray Freddie. International Journal of Cancer.2014;136(5). CrossRef
- Cancer statistics, 2013 Siegel Rebecca, Naishadham Deepa, Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2013;63(1). CrossRef
- Oral squamous cell carcinoma: Review of prognostic and predictive factors Massano João, Regateiro Frederico S., Januário Gustavo, Ferreira Artur. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology.2006;102(1). CrossRef
- Salivary zinc finger protein 510 peptide as a novel biomarker for detection of oral squamous cell carcinoma in early stages Jou Yu-Jen, Lin Chia-Der, Lai Chih-Ho, Tang Chih-Hsin, Huang Su-Hua, Tsai Ming-Hsui, Chen Shih-Yin, Kao Jung-Yie, Lin Cheng-Wen. Clinica Chimica Acta.2011;412(15-16). CrossRef
- Perioperative Complications, Comorbidities, and Survival in Oral or Oropharyngeal Cancer Ribeiro Karina de Cássia Braga, Kowalski Luiz Paulo, Latorre Maria do Rosário Dias de Oliveira. Archives of Otolaryngology–Head & Neck Surgery.2003;129(2). CrossRef
- The different roles of ER subtypes in cancer biology and therapy Thomas Christoforos, Gustafsson Jan-Åke. Nature Reviews Cancer.2011;11(8). CrossRef
- Application of a multigene reverse transcription-PCR assay for detection of mammaglobin and complementary transcribed genes in breast cancer lymph nodes Zehentner BK, Dillon DC , Jiang Y , Xu J, Bennington A, Molesh DA, et al . Clinical chemistry.2002;48(8):1225-1231.
- MicroRNAs: Novel regulators in the hallmarks of human cancer Ruan Kai, Fang Xiaoguang, Ouyang Gaoliang. Cancer Letters.2009;285(2). CrossRef
- A role for microRNAs in the development of the immune system and in the pathogenesis of cancer Kanellopoulou Chryssa, Monticelli Silvia. Seminars in Cancer Biology.2008;18(2). CrossRef
- MicroRNA therapeutics in preclinical cancer models Kim Minlee, Kasinski Andrea L, Slack Frank J. The Lancet Oncology.2011;12(4). CrossRef
- MicroRNAs: miRRORS of health and disease Montano Monty. Translational Research.2011;157(4). CrossRef
- MicroRNAs and head and neck cancer: Reviewing the first decade of research Sethi Neeraj, Wright Alexander, Wood Henry, Rabbitts Pamela. European Journal of Cancer.2014;50(15). CrossRef
- The role of microRNAs in cancer: No small matter Wiemer Erik A.C.. European Journal of Cancer.2007;43(10). CrossRef
-
Localization and Concentration of Carcinoembryonic Antigen (CEA) in Gastrointestinal Tumors: Correlation With CEA Levels in Plasma
2 JNCI: Journal of the National Cancer Institute.1981. CrossRef - Molecular themes in oncogenesis Bishop J.Michael. Cell.1991;64(2). CrossRef
- MiR-21 Indicates Poor Prognosis in Tongue Squamous Cell Carcinomas as an Apoptosis Inhibitor Li J., Huang H., Sun L., Yang M., Pan C., Chen W., Wu D., Lin Z., Zeng C., Yao Y., Zhang P., Song E.. Clinical Cancer Research.2009;15(12). CrossRef
- MicroRNA Expression Ratio Is Predictive of Head and Neck Squamous Cell Carcinoma Avissar M., Christensen B. C., Kelsey K. T., Marsit C. J.. Clinical Cancer Research.2009;15(8). CrossRef
- Real- time PCR: current technology and applications: Horizon Scientific Press; 2009 Logan J , Logan JM , Edwards KJ , Saunders NA. .
- MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients Zheng Yuanyuan, Cui Long, Sun Weiliang, Zhou Hui, Yuan Xianbin, Huo Ming, Chen Jie, Lou Yanru, Guo Junming. Cancer Biomarkers.2012;10(2). CrossRef
- Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue Wong T.-S., Liu X.-B., Wong B. Y.-H., Ng R. W.-M., Yuen A. P.-W., Wei W. I.. Clinical Cancer Research.2008;14(9). CrossRef
- miR-24 up-regulation in oral carcinoma: Positive association from clinical and in vitro analysis Lin Shu-Chun, Liu Chung-Ji, Lin Jung-An, Chiang Wei-Fan, Hung Pei-Shih, Chang Kuo-Wei. Oral Oncology.2010;46(3). CrossRef
- Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma Liu Chung-Ji, Lin Shu-Chun, Yang Cheng-Chieh, Cheng Hui-Wen, Chang Kuo-Wei. Head & Neck.2011;34(2). CrossRef
- CA15.3, CEA and TPA Tumor Markers in the Early Diagnosis of Breast Cancer Relapse Vizcarra E., Lluch A., Cibrián R., Jarque F., García-Conde J.. Oncology.1994;51(6). CrossRef
- Pre-operative pulmonary rehabilitation and surgery for lung cancer Cesario Alfredo, Ferri Luigi, Galetta Domenico, Cardaci Vittorio, Biscione Gianluca, Pasqua Franco, Piraino Alessio, Bonassi Stefano, Russo Patrizia, Sterzi Silvia, Margaritora Stefano, Granone Pierluigi. Lung Cancer.2007;57(1). CrossRef
- Detection of CK19 and CEA mRNA expression for the diagnosis of peripheral blood micrometastases in patients with non-small cell lung cancer Zhu G, Liu D, Wang X, Chen J. Zhongguo Fei Ai Za Zhi.2004;7(3):226-229.
- MicroRNA Gene Expression Deregulation in Human Breast Cancer Iorio Marilena V., Ferracin Manuela, Liu Chang-Gong, Veronese Angelo, Spizzo Riccardo, Sabbioni Silvia, Magri Eros, Pedriali Massimo, Fabbri Muller, Campiglio Manuela, Ménard Sylvie, Palazzo Juan P., Rosenberg Anne, Musiani Piero, Volinia Stefano, Nenci Italo, Calin George A., Querzoli Patrizia, Negrini Massimo, Croce Carlo M.. Cancer Research.2005;65(16). CrossRef
- Analysis of miR-205 and miR-155 expression in the blood of breast cancer patients Liu J, Mao Q, Liu Y, Hao X, Zhang S, Zhang J. Chin J Cancer Res .2013;25(1):46-54.
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2020
Author Details