CDKs family -a glimpse into the past and present: from cell cycle control to current biological functions
Download
Abstract
Cyclin-dependent kinases (CDKs) are the catalytic subunits or protein kinases characterized by separate subunit “cyclin” that are essential for their enzymatic activity. CDKs play important roles in the control of cell cycle progression, cell division, neuronal function, epigenetic regulation, metabolism, stem cell renewal and transcription. However, they can accomplish some of these tasks independently, without binding with cyclin protein or kinase activity. Thus, so far, twenty different CDKs and cyclins have been reported in mammalian cells. The evolutionary expansion of the CDK family in mammals led to the division of CDKs into three cell-cycle-related subfamilies (Cdk1, Cdk4 and Cdk5) and five transcriptional subfamilies (Cdk7, Cdk8, Cdk9, Cdk11 and Cdk20). In this review, we summarizes that how CDKs are traditionally involve their latest revelations, their functional diversity beyond cell cycle regulation and their impact on development of disease in mammals.
Introduction
The cellular processes such as cell cycle is driven by protein kinases referred to as “Cyclin dependent kinases” (CDKs) whose serine/threonine-specific catalytic core, control the kinase activity and are only activated when bound by specific regulatory subunit “cyclin”. This CDKs activity is regulated by phosphorylation of a target protein through CDK’s T-loop and binding of inhibitory proteins [1].
CDKs were first discovered through biological and genetic studies in yeast [2-5]. In human, there are twenty distinct family members of CDKs have been described, which have been involved in two main process transcription and cell division between distinct phases of the cell cycle through specific substrate phosphorylation [6-7].
In this review, we first summarize the most relevant information for known CDKs, with a particular emphasis on those involved in regulating the cell cycle. We then discuss other observations derived from biological studies based on animals and human models.
In 1987 first human kinases, CDK1 was cloned by using functional complementation in yeast, and was termed cell division cycle 2 (Cdc2) because of its high homology with fission yeast kinase Cdc2. CDK1 bind to cyclin A and B and encoded by the Cdc2 gene [2], these complexes drive the transition between G2 phase and M phase, as well as early M phase.
Discussion
In higher eukaryotes, CDK1 and CDK2 emerged as key determinant of mitotic progression and DNA replication respectively. However, they regulate the G1/S and G2/M phases of the cell cycle by binding with cyclin E or A and cyclin B kinase, respectively [8]. Cyclin E binds G1 phase Cdk2, which is required for the transition from G1 to S phase while binding with Cyclin A is required to progress through the DNA synthetic S phase (Figure 1).
Figure 1: Function of CDKs and Cyclin (CDK/Cyclin) Complexes at Specific Phases of the Cell Cycle..
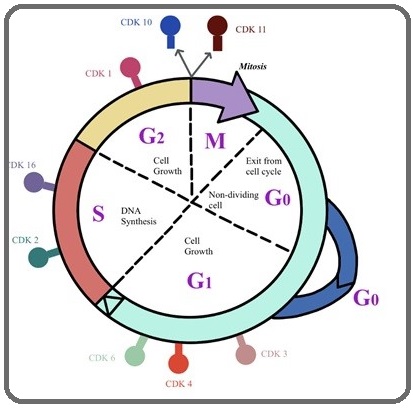
Previous study has revealed biological role of CDK2 in cellular proliferation, cell death, and DNA repair in human embryonic stem cells (HESC) [9]. A recent study by Mori et al [10] showed Cep169, a centrosomal protein conserved among vertebrates, dissociation is controlled by Cdk1/Cyclin B during mitosis.
CDK3 is the closest relative to CDK2 among mammalian CDK genes identified thus far and has originally been classified as a cyclin dependent kinase, because of its high sequence identity with CDK2 and the ability to complement cdc28 mutations in yeast [11]. It executes an essential function at the G1/S transition (Figure 1) in the mammalian cell cycle (van den Heuvel 1993). CDK3 binds with Cyclin C and regulate the Rb-dependent G0/G1 transition [12] while enhancing the transactivation and transcriptional activities of the transcription factor 1 (TF1) by phosphorylation [13].
Previously it was revealed that CDK4 and CDK6 are dispensable for cell cycle progression and are essential for development and differentiation of highly specialized cell types [14]. However, recently Sherr CJ and his group reported their roles in mammalian cell proliferation, where they help to drive the progression of cells into the S phase of the cell division cycle (Figure 1) [15]. Through association and activation of CDK4 and CDK6 with D-Type cyclins, promotes progression to G1 phase, however, CDK4 inhibition has been shown to induce G1 arrest and apoptosis [16-17].
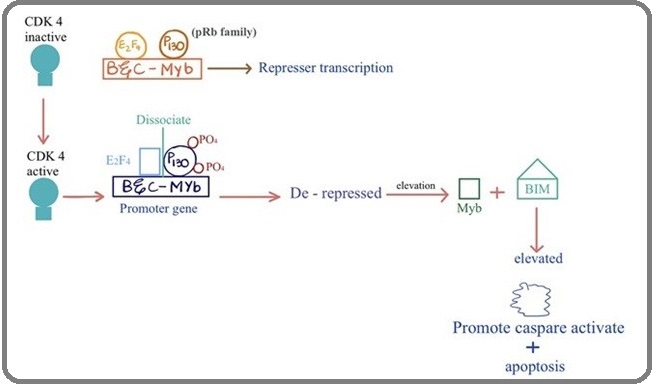
In addition, CDK4 and CDK6 are also involved in promoting cell death in neurons during development and disease. CDK4 has been known in the regulation of neuronal cell death, while activation of CDK4 leads to hyper-phosphorylation of the pRb family member p130, dissociation of p130 and associated chromatin modifiers from the transcription factor E2F4. However, pro-apoptotic BH3-only protein Bim, (Bcl-2-like protein 11) is stimulated by expression of E2F binding genes including the transcription factors B- and C-Myb (myeloblastosis) [18]. Previously it was also reported that deregulation of CDK4 and CDK6 kinase with cyclin D resulting in Rb hyperphosphorylation associated with a loss of control between mitogenic stimuli and cell cycle regulation, which leads to uncontrolled cell proliferation and apoptosis (Figure 2) [19].
Figure 2: CDK4 Involvement in Neuronal Cell Death.
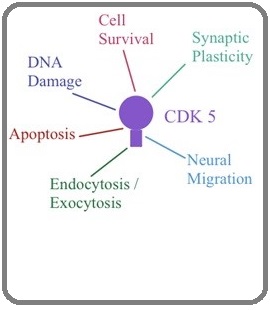
CDK5 is unusual because it is not believed to be active in a typical cell cycle while it binds to cyclin protein. It is well characterized for its role in the central nervous system, terminally differentiated and proliferating cells rather than in the cell cycle [20]. Recent study showed Cdk5-ATM (ataxia-telangiectasia mutated) pathway plays a crucial role in DNA damage-induced neuronal injury [21]. It was previously reported that Cdk5 retards closure of an in vitro scrape wound in a mouse corneal epithelial cell line and strengthens cell-matrix adhesion and possible biological function of CDK5 described in Figure 3 [22].
Figure 3: Diverse Biological Function of CDK5..
Subsequently, CDK7, CDK8 and CDK9 were identified and are known to directly promote the cell cycle and regulate the transcription [23-25]. CDK7 associates with Cyclin H and forms a complex termed CAK, the CDK-Activating Kinase, this complex phosphorylate cell-cycle CDKs within the activation segment (T-loop), and also a component of the general transcription factor TFIIH, which phosphorylates the C-terminal domain (CTD) of Pol II [26] (Figure 4a)(Figure 4).
Figure 4: Biological Activity of CDKs (a) CDK7, (b and c) CDK8, (d) CDK9, (e) CDK10.
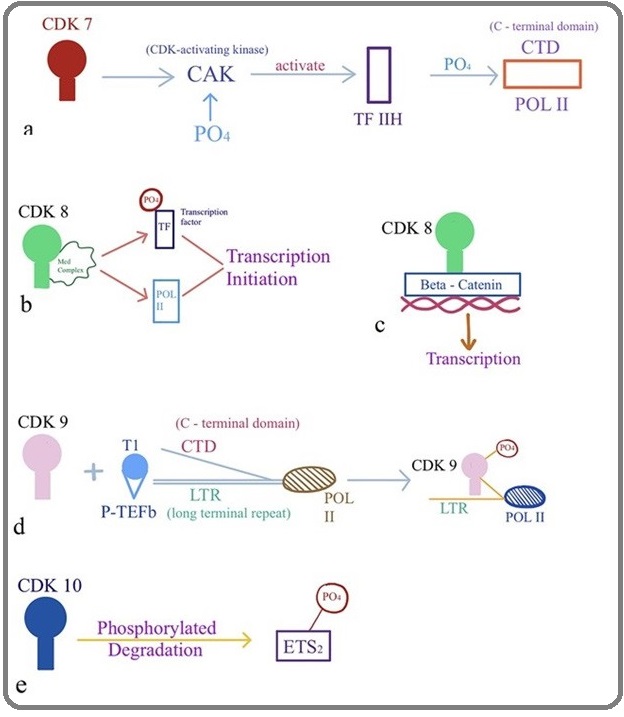
Another function of CDK7 emerged in neocortical development and proper expression levels of both CDK7 and miR-210 are required for normal Neural Progenitors cell-cycle progression [27]. In addition, CDK8 as part of mediator complex, regulates gene expression through phosphorylation of transcription factors [28]. Moreover, this complex controls the Mediator–pol II interaction to help in the transcription initiation and reinitiating events which are required for expression of protein-coding genes, this may reflect a common mechanism in the human cells by which activated transcription is shut down [29] (Figure 4b). Moreover, it is also required for cell division associated with Wnt/ β-catenin signaling (Figure 4c), [30-31] and act as a novel regulator of p27 by facilitating Skp2 (S-phase kinase-associated protein 2)-mediated ubiquitination and degradation of p27 in breast cancer [32] .
Cyclin T1, T2a, T2b, or K associates with CDK9 to form active positive transcription elongation factor (P-TEFb) complexes, resulting in activation of the transcriptional elongation by phosphorylating the C-terminal domain (CTD) of RNA polymerase II (Figure 4 d) (RNAPII) [33-34]. Previously it has also been reported that CDK9 predominantly involved in co-transcriptional histone modification, messenger RNA (mRNA) processing,mRNA export and DNA repair [35].
CDK10 was discovered by sequence homology screening for CDK-related genes and plays a role in the cell cycle through acting during the G2 or M phase (Figure 1) [36]. Currently, it was reported that it acts as the regulator of the ETS2 transcription factor and modulates its transactivation activity (Figure 4 e) [37]. However, for the past twenty years and until recently, the elucidation of the functions of CDK10 was hampered by the lack of any identified cyclin partner. Guen etal has reported siRNA mediated silencing of cyclin M causes extreme reduction of CDK10 expression in human cells [38]. In addition, several studies have shown reduced expression of CDK10 in many cancer, demonstrating its putative role as tumor suppressor gene in multiple types of human cancers [39-42].
CDK11 binds with cyclins L and has role in transcription, RNA processing in particular alternative splicing [43-45]. It is also participates in many other pathways, such as hormone receptor signaling or autophagy [46-48]. Various studies have demonstrated that Cdk11, is specifically expressed at G2-M, (Figure 1) and during mitosis its kinase activity is required for duplication of the ntrioles, spindle dynamics and sister chromatid cohesion at centromeres [44,49-50].
The Cdk12 and Cdk13, both paired with cyclin K and identified in cDNA screens for cell cycle regulators. They were initially named CRKRS and CDC2L5 [51] and play role in regulation of transcription through the differential phosphorylation of the C-terminal domain (CTD) of RNA Polymerase II [7-52-53].
CDK12 (alias CRKRS, CRK7, CRKR, KIAA0904) was originally identified as a Cdc2-related serine/threonine kinase (STK) possessing an arginine/serine (RS)-rich domain, which was closely related to the family of CDKs. [52-54]. Previously Chen and his group have reported its interaction with cyclins L1 and L2 (CycL) [55], however various studies have now identified cyclin K (CycK), as the endogenous binding CDK12 partner [53-56].
Ko et al. proposed that CDK12 could play a role in the regulation of transcription and alternative splicing rather than cell cycle progression [51]. They have hypothesized that CDK12 could be a novel RNA polymerase II (RNAPII) kinase that might directly link transcription with the splicing machinery. Previously Rodrigues et al. proposed that CDK 12 acts a splicing regulator (Figure 5a) for glial-specific splicing of NeurexinIV on specific pre-mRNA sites defined by HOW (sequence specific RNA binding protein) [57]. CDK12 has also shown an indirect role in the cellular process of DNA damage response (DDR) and maintenance of genomic stability by modulating the expression of DDR genes. The authors also demonstrated that CycK/CDK12 depletion increases the number of cells in the G2-M phase of the cell cycle [53].
Figure 5: Biological role of CDKs (a) CDK12, (b) CDK13, (c) CDK14, (d) CDK19, (e) CDK20.
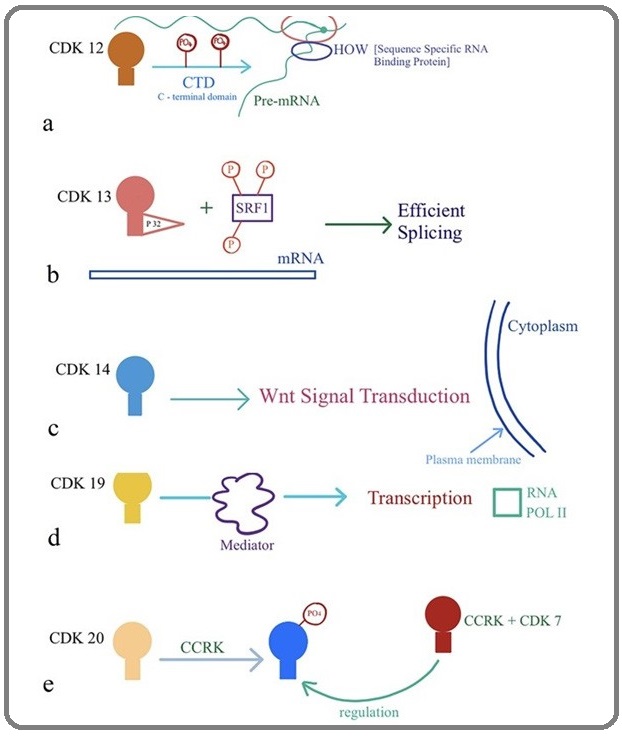
CDK13 protein kinase is also involved in the regulation of gene expression by controlling the phosphorylation status and activity of splicing regulators [54-58]. It is part of a family of 20 different ATP-dependent serine-threonine protein kinases regulating cell-cycle progression and gene expression [6]. It is known to interact with two types of regulatory subunits, K and L-type cyclins [53-55]. In addition, CDK13 interacts with p32 a protein associating with the splicing factor SRSF1 (also known as ASF/SF2) and by phosphorylating SRSF1 (Figure 5b), this complex increases the mRNA splicing of human immunodeficient virus type 1 (HIV-1) while its overexpression, suppresses virus production [59].
The activity of some CDKs requires protein motif PFTAIRE (Cdc2-related kinases) which mediates binding to co-activating proteins called cyclins and has been classify other newly identified CDKs including CDK14 (PFTK1), CDK15 (PFTK2), CDK16 (PCTK1),CDK17 (PCTK2) and CDK18 (PCTK3) or on a sequence homology with the CDKs, such as CDC2-like kinase (CDK19) or cell cycle-related kinase (CDK20) [60]. Previously it was reported that CDK14 associated with cyclin Y and exert their influence over Wnt signal transduction (Figure 5 c) remotely at the cell surface which are anchored to the plasma membrane [61-62]. Furthermore, CDk14 over expression has been found in various human cancers [63-65].
The PFTK2/CDK15 is very poorly characterized kinases, and little is known about its expression and regulation. Evolutionarily, CDK15 seems to be of a newer origin, which is more similar to CDK14 (PFTK1). APrevious study found that PCTK-1/CDK16 is present in the cytoplasm throughout the cell cycle and displays kinase activity during S phase and the G2 phase (Figure 1) and correlated with dephosphorylation of tyrosine residues. [66]. Abundant expression of Cdk16 was also detected in post-mitotic brain cells [67] and subsequently, high levels of CDK16 are found in the cytoplasm of cerebellar Purkinje cells, as well as in cells of the hippocampus and the neocortex [68]. In mammals, CDK16 is required for spermatogenesis, [69] polarization of presynaptic vesicles and synapse elimination during neural circuit rewiring in nematodes [70-71].
Hirose T et al and his group found transcripts of rat PCTK2/CDK17 in the hippocampal and olfactory bulb regions of the brain [72]. It was also shown to interact with TRAP (Tudor repeat associated with PCTK2)16 as well as cables (adaptor molecule linking the non-receptor tyrosine kinase c-abl with CDKs) [73].
PCTAIRE kinase 3 (PCTK3) or CDK18 was first reported in human Alzheimer’s brain as neuronal kinase that phosphorylates TAU protein [74]. Previous study showed the mechanisms of catalytic activation of PCTK3 by cyclin A2 and protein kinase [75]. It was also showed that cdk18 has role in replication stress signaling and serves as a novel regulator of genome integrity [76].
The cdk19 (previously known as CDK8-like, CDK8L or CDC2L6) protein is similar to cdk8, although both CDK8 and CDK19 associate with C type cyclin as a part of the multi-subunit Mediator complexes [4,6-77-78] which links transcription factors with Pol II [79]. However, a recent study identified a novel links between CDK19 and cell proliferation, p53 response, and cholesterol metabolism [80]. Established and emerging functions of CDKs are summarized in Table 1.
| Protein | Cyclin binding element | Cyclin | Kinase Activity | Cellular Function | References |
| CDK1 | PSTAIRE | A & B | Yes | Control G2 & M Phase,FoxM1 and FoxK2 transcription in complex with cyclin B,ESC self-renewal through interaction with Oct4, NSC self-renewal through inhibition of Ngn2, HR-mediated DNA damage repair, Epigenetic regulation through Ezh2 and Dnmt1, dissociation of Cep169 from centrosomes is controlled by Cdk1/Cyclin B during mitosis | (10, 36, 55) |
| CDK2 | PSTAIRE | E & A | Yes | Control of G1-S Phase of cell cycle Promote S phase entry by USP37 activation Myoblast proliferation through inhibition of MyoD Rb/E2F transcription FoxM1 and FoxK2 transcription in complex with cycA NSC self-renewal through inhibition of Ngn2 Epigenetic regulation through Ezh2 and Dnmt1 | (55, 83) |
| CDK3 | PSTAIRE | C | NHEJ-mediated DNA damage repair in complex with cyclin C | (14, 15, 84) | |
| CDK4 | PISTVRE | D | Yes | Control G 1 Phase of cell cycle, Rb/E2F transcription Epigenetic regulation through Mep50 | |
| CDK5 | PSSALRE | None | Yes | Activated by non-cyclin proteins, including Cdk5R1 (p35) and Cdk5R2 (p39), Neuronal function in complex with p35 and p39, Epigenetic regulation through Dnmt1, Glycogen synthesis Strengthens cell-matrix adhesion and retards closure of an in vitro scrape wound in a mouse corneal epithelial cell line | (22) |
| CDK6 | PLSTIRE | D | Yes | Control of G1 Phase of cell cycle; Rb/E2F Transcription progression of cells into the DNA synthetic (S) phase | |
| CDK7 | NRTALRE | H | Yes | Cdk-activating kinase (CAK) and RNAPII transcription in complex with cyclin H | (26) |
| CDK8 | SMSACRE | C | Yes | G1 & G2 Phase of cell cycle RNAP II transcription in complex with Cyclin C, Wnt/β-catenin pathway in complex with cyclin C, Inhibition of lipogenesis in complex with cyclin C | (29, 31) |
| CDK9 | PITALRE | T1 T2a T2 b K | Yes | RNAPII transcription in complex with Cyclin T, DNA damage response in complex with cyclin K cdk9-cyclin k in maintaining genome integrity | (33, 34) |
| CDK10 | PISSLRE | M | Yes | G2/M Phase Ets2 transcription | (36, 38) |
| CDK11 | PITSLRE | L | Yes | G2/M Phase RNA splicing in complex with cyclin L | (43-45) |
| CDK12 | PITAIRE | K/L | Yes | RNAPII transcription in complex with cyclin K DNA damage response in complex with cyclin K | (7, 53, 56) |
| CDK13 | PITAIRE | K/L | Yes | RNAPII transcription in complex with cyclin K | (7, 53, 56) |
| CDK14 | PITAIRE | Y | Yes | Wnt/ β-catenin pathway in complex with cyclin Y | (61, 62) |
| CDK15 | PITAIRE | Y | Yes | Synaptic trafficking and remodeling in complex with cyclin Y | (70, 71) |
| CDK16 | PCTAIRE | Y | Yes | PCTAIRE proteins or PCTK1/ displays kinase activity during S phase and the G2 phase/ Spermatogenesis in complex with Cyclin Y | (66, 69, 73) |
| CDK17 | PCTAIRE | Y | Yes | PCTAIRE proteins or PCTK2/ iSer/Thr kinase that might play a unique role in terminally differentiated neurons. | (72) |
| CDK18 | PCTAIRE | K | Yes | post-mitotic Function PCTAIRE proteins or PCTK3/ phosphorylates TAU protein regulator of genome integrity | (60, 74, 76) |
| CDK19 | SMSACRE | C | Associated with C-type cyclins as part of the multi-subunit Mediator complex Links to transcription factors with Pol II | (60, 74, 76, 77, 79) | |
| CDK20 | PNQALRE | H | Yes | CAK (CDK-activating kinase) activity for Cdk2, activating kinase for MAK-related kinase/intestinal cell kinase (ICK) activates β-catenin-TCF signaling to stimulate cell-cycle progression | (81, 82) |
Finally, Cdk20 (also known as cell cycle-related kinase (CCRK), is associated with cyclin H and known as an important regulator of G1- to S-phase transition in cell cycle while it has CDK activating kinase (CAK) activity for Cdk2, suggesting a close relationship with Cdk7 [81]. Expression of Cdk20 causes activation of β-catenin-TCF signaling which in turn to stimulate the cell-cycle progression [82], whereas CAK inhibition results in accumulation of intestinal cell kinases at the ciliary tips and prevents cell-cycle entry [65].
Thus far, CDKs family implicated in transcription, DNA damage repair, proteolytic degradation, epigenetic regulation, and metabolism, stem cell self-renewal, neuronal functions and spermatogenesis.
In conclusions, CDKs and multifaceted proteins cyclins are the essential regulators of the cell cycle and have a tremendous role in different biological processes that are distinct from cell division. However, the majority of these emerging functions are closely intertwined with the cell cycle.
Abbreviations
CDK: Cyclin dependent Kinases CDC2: Cell division cycle 2
HESC: human embryonic stem cells
CTD: C Terminal Domain
TF1: Transcription factor 1
ATM: ataxia-telangiectasia mutated CCRK: Cell cycle-related kinase
SKP2: S-phase kinase associated protein Pol II: Polymerase II
P-TEF: Positive Transcription Elongation factor
Competing interests
The authors declare that they have no competing interests
Authors’ contributions
SM designed the basic frame work and outline of manuscript, wrote and revised manuscript. MS, MFHQ, DM, ML and TU designed all the graphics, managed literature searches and provided help in manuscript preparation. All authors have read and agreed to the published version of manuscript.
Acknowledgements
Not applicable
References
- Molecular Cell Biology. 4th edition. ed. New York: W. H. Freeman; 2000 Lodish H BA, Zipursky SL, et al . .
- Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2 Lee Melanie G., Nurse Paul. Nature.1987;327(6117). CrossRef
- NOBEL LECTURE: Cyclin Dependent Kinases and Cell Cycle Control Nurse Paul M.. Bioscience Reports.2002;22(5-6). CrossRef
- The Protein Kinase Complement of the Human Genome Manning G.. Science.2002;298(5600). CrossRef
- Mammalian cyclin-dependent kinases Malumbres Marcos, Barbacid Mariano. Trends in Biochemical Sciences.2005;30(11). CrossRef
- Cyclin-dependent kinases: a family portrait Malumbres Marcos, Harlow Edward, Hunt Tim, Hunter Tony, Lahti Jill M., Manning Gerard, Morgan David O., Tsai Li-Huei, Wolgemuth Debra J.. Nature Cell Biology.2009;11(11). CrossRef
- Interaction of Cyclin-Dependent Kinase 12/CrkRS with Cyclin K1 Is Required for the Phosphorylation of the C-Terminal Domain of RNA Polymerase II Cheng S.- W. G., Kuzyk M. A., Moradian A., Ichu T.-A., Chang V. C.- D., Tien J. F., Vollett S. E., Griffith M., Marra M. A., Morin G. B.. Molecular and Cellular Biology.2012;32(22). CrossRef
- Four-dimensional control of the cell cycle Pines Jonathon. Nature Cell Biology.1999;1(3). CrossRef
- An Important Role for CDK2 in G1 to S Checkpoint Activation and DNA Damage Response in Human Embryonic Stem Cells Neganova Irina, Vilella Felipe, Atkinson Stuart P., Lloret Maria, Passos João F., von Zglinicki Thomas, O'Connor José-Enrique, Burks Deborah, Jones Richard, Armstrong Lyle, Lako Majlinda. STEM CELLS.2011;29(4). CrossRef
- Phosphorylation of the centrosomal protein, Cep169, by Cdk1 promotes its dissociation from centrosomes in mitosis Mori Yusuke, Inoue Yoko, Taniyama Yuki, Tanaka Sayori, Terada Yasuhiko. Biochemical and Biophysical Research Communications.2015;468(4). CrossRef
- A family of human cdc2-related protein kinases Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, Nelson C, et al . The EMBO journal.1992;11(8):2909-2917.
- Cyclin C/Cdk3 Promotes Rb-Dependent G0 Exit Ren Shengjun, Rollins Barrett J. Cell.2004;117(2). CrossRef
- Cyclin-Dependent Kinase 3-Mediated Activating Transcription Factor 1 Phosphorylation Enhances Cell Transformation Zheng D., Cho Y.-Y., Lau A. T.Y., Zhang J., Ma W.-Y., Bode A. M., Dong Z.. Cancer Research.2008;68(18). CrossRef
- Living with or without cyclins and cyclin-dependent kinases Sherr C. J.. Genes & Development.2004;18(22). CrossRef
- Targeting CDK4 and CDK6: From Discovery to Therapy Sherr C. J., Beach D., Shapiro G. I.. Cancer Discovery.2015;6(4). CrossRef
- Signaling through cyclin D-dependent kinases Choi Y J, Anders L. Oncogene.2013;33(15). CrossRef
- A possible usage of a CDK4 inhibitor for breast cancer stem cell-targeted therapy Han Yu Kyeong, Lee Jae Ho, Park Ga-Young, Chun Sung Hak, Han Jeong Yun, Kim Sung Dae, Lee Janet, Lee Chang-Woo, Yang Kwangmo, Lee Chang Geun. Biochemical and Biophysical Research Communications.2013;430(4). CrossRef
- Cell cycle molecules define a pathway required for neuron death in development and disease Greene Lloyd A., Liu David X., Troy Carol M., Biswas Subhas C.. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease.2007;1772(4). CrossRef
- A revised picture of the E2F transcriptional network and RB function Stevaux Olivier, Dyson Nicholas J. Current Opinion in Cell Biology.2002;14(6). CrossRef
- Cyclin-dependent kinases Malumbres M. Genome Biology.2014;15(6):122.
- Study of ATM Phosphorylation by Cdk5 in Neuronal Cells She H, Mao Z. Methods in molecular biology (Clifton, NJ).2017;1599:363-374.
- CDK5 regulates cell adhesion and migration in corneal epithelial cells Gao C, Negash S, Guo HT, Ledee D, Wang HS, Zelenka P. Molecular cancer research : MCR.2002;1(1):12-24.
- The history and future of targeting cyclin-dependent kinases in cancer therapy Asghar Uzma, Witkiewicz Agnieszka K., Turner Nicholas C., Knudsen Erik S.. Nature Reviews Drug Discovery.2015;14(2). CrossRef
- Cdks, cyclins and CKIs: roles beyond cell cycle regulation Lim S., Kaldis P.. Development.2013;140(15). CrossRef
- The two faces of Cdk8, a positive/negative regulator of transcription Nemet Josipa, Jelicic Branka, Rubelj Ivica, Sopta Mary. Biochimie.2014;97. CrossRef
- The role of Cdk7 in CAK function, a retro-retrospective Harper J. W., Elledge S. J.. Genes & Development.1998;12(3). CrossRef
- CDK7 and miR-210 Co-regulate Cell-Cycle Progression of Neural Progenitors in the Developing Neocortex Abdullah Aisha I., Zhang Haijun, Nie Yanzhen, Tang Wei, Sun Tao. Stem Cell Reports.2016;7(1). CrossRef
- The human Mediator complex: a versatile, genome-wide regulator of transcription Taatjes Dylan J.. Trends in Biochemical Sciences.2010;35(6). CrossRef
- Emerging roles of Cdk8 in cell cycle control Szilagyi Zsolt, Gustafsson Claes M.. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms.2013;1829(9). CrossRef
- CDK8 Is a Stimulus-Specific Positive Coregulator of p53 Target Genes Donner Aaron Joseph, Szostek Stephanie, Hoover Jennifer Michelle, Espinosa Joaquin Maximiliano. Molecular Cell.2007;27(1). CrossRef
- CDK8 regulates E2F1 transcriptional activity through S375 phosphorylation Zhao J, Ramos R, Demma M. Oncogene.2012;32(30). CrossRef
- Skp2–MacroH2A1–CDK8 axis orchestrates G2/M transition and tumorigenesis Xu Dazhi, Li Chien-Feng, Zhang Xian, Gong Zhaohui, Chan Chia-Hsin, Lee Szu-Wei, Jin Guoxiang, Rezaeian Abdol-Hossein, Han Fei, Wang Jing, Yang Wei-Lei, Feng Zi-Zhen, Chen Wei, Wu Ching-Yuan, Wang Ying-Jan, Chow Lu-Ping, Zhu Xiao-Feng, Zeng Yi-Xin, Lin Hui-Kuan. Nature Communications.2015;6(1). CrossRef
- The multi-tasking P-TEFb complex Brès Vanessa, Yoh Sunnie M, Jones Katherine A. Current Opinion in Cell Biology.2008;20(3). CrossRef
- P-TEFb, a Cyclin-Dependent Kinase Controlling Elongation by RNA Polymerase II Price David H.. Molecular and Cellular Biology.2000;20(8). CrossRef
- CDK9 directs H2B monoubiquitination and controls replication‐dependent histone mRNA 3′‐end processing Pirngruber Judith, Shchebet Andrei, Schreiber Lisa, Shema Efrat, Minsky Neri, Chapman Rob D, Eick Dirk, Aylon Yael, Oren Moshe, Johnsen Steven A. EMBO reports.2009;10(8). CrossRef
- The cdc2-related Kinase, PISSLRE, Is Essential for Cell Growth and Acts in G2 Phase of the Cell Cycle Li S, MacLachlan TK, De Luca A, Claudio PP, Condorelli G, Giordano A. 1995;55(18):3992-3995.
- The awakening of the CDK10/Cyclin M protein kinase Guen Vincent J., Gamble Carly, Lees Jacqueline A., Colas Pierre. Oncotarget.2017;8(30). CrossRef
- CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome Guen V. J., Gamble C., Flajolet M., Unger S., Thollet A., Ferandin Y., Superti-Furga A., Cohen P. A., Meijer L., Colas P.. Proceedings of the National Academy of Sciences.2013;110(48). CrossRef
- ThePISSLREGene: Structure, Exon Skipping, and Exclusion as Tumor Suppressor in Breast Cancer Crawford J, Ianzano L, Savino M, Whitmore S, Cleton-Jansen A-M, Settasatian C, et al . Genomics .1999;56(1):90-97.
- Identification of CDK10 as an Important Determinant of Resistance to Endocrine Therapy for Breast Cancer Iorns Elizabeth, Turner Nicholas C., Elliott Richard, Syed Nelofer, Garrone Ornella, Gasco Milena, Tutt Andrew N.J., Crook Tim, Lord Christopher J., Ashworth Alan. Cancer Cell.2008;13(2). CrossRef
- Clinical and biological significance of Cdk10 in hepatocellular carcinoma Zhong Xiang-yu, Xu Xiao-xue, Yu Jian-hua, Jiang Gui-xing, Yu Yang, Tai Sheng, Wang Zhi-dong, Cui Yun-fu. Gene.2012;498(1). CrossRef
- CDK10 functions as a tumor suppressor gene and regulates survivability of biliary tract cancer cells YU JIAN-HUA, ZHONG XIANG-YU, ZHANG WEI-GUANG, WANG ZHI-DONG, DONG QIN, TAI SHENG, LI HUI, CUI YUN-FU. Oncology Reports.2011;27(4). CrossRef
- Characterization of Cyclin L1 and L2 Interactions with CDK11 and Splicing Factors Loyer Pascal, Trembley Janeen H., Grenet Jose A., Busson Adeline, Corlu Anne, Zhao Wei, Kocak Mehmet, Kidd Vincent J., Lahti Jill M.. Journal of Biological Chemistry.2008;283(12). CrossRef
- CDK11p58 is required for the maintenance of sister chromatid cohesion Hu D., Valentine M., Kidd V. J., Lahti J. M.. Journal of Cell Science.2007;120(14). CrossRef
- CDK11 Complexes Promote Pre-mRNA Splicing Hu Dongli, Mayeda Akila, Trembley Janeen H., Lahti Jill M., Kidd Vincent J.. Journal of Biological Chemistry.2002;278(10). CrossRef
- Polyubiquitination inhibition of estrogen receptor alpha and its implications in breast cancer Tecalco-Cruz Angeles C, Ramírez-Jarquín Josué O. World Journal of Clinical Oncology.2018;9(4). CrossRef
- CDK11p58 represses vitamin D receptor-mediated transcriptional activation through promoting its ubiquitin–proteasome degradation Chi Yayun, Hong Yi, Zong Hongliang, Wang Yanlin, Zou Weiying, Yang Junwu, Kong Xiangfei, Yun Xiaojing, Gu Jianxin. Biochemical and Biophysical Research Communications.2009;386(3). CrossRef
- The cyclin-dependent kinase PITSLRE/CDK11 is required for successful autophagy Wilkinson Simon, Croft Daniel R., O’Prey Jim, Meedendorp Arenda, O’Prey Margaret, Dufès Christine, Ryan Kevin M.. Autophagy.2011;7(11). CrossRef
- The PITSLRE/CDK11 p58 protein kinase promotes centrosome maturation and bipolar spindle formation Petretti Clotilde, Savoian Matthew, Montembault Emilie, Glover David M, Prigent Claude, Giet Régis. EMBO reports.2006;7(4). CrossRef
- Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate Yokoyama Hideki, Gruss Oliver J., Rybina Sofia, Caudron Maïwen, Schelder Malgorzata, Wilm Matthias, Mattaj Iain W., Karsenti Eric. Journal of Cell Biology.2008;180(5). CrossRef
- CrkRS: a novel conserved Cdc2- related protein kinase that colocalises with SC35 speckles Ko TK, Kelly E, Pines J. Journal of cell science. 2001;114():.2001;114(Pt 14):2591-2603.
- Structural and Functional Analysis of the Cdk13/Cyclin K Complex Greifenberg Ann Katrin, Hönig Dana, Pilarova Kveta, Düster Robert, Bartholomeeusen Koen, Bösken Christian A., Anand Kanchan, Blazek Dalibor, Geyer Matthias. Cell Reports.2016;14(2). CrossRef
- The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B. M.. Genes & Development.2011;25(20). CrossRef
- Characterization of Human Cyclin-Dependent Kinase 12 (CDK12) and CDK13 Complexes in C-Terminal Domain Phosphorylation, Gene Transcription, and RNA Processing Liang Kaiwei, Gao Xin, Gilmore Joshua M., Florens Laurence, Washburn Michael P., Smith Edwin, Shilatifard Ali. Molecular and Cellular Biology.2015;35(6). CrossRef
- Identification and Characterization of the CDK12/Cyclin L1 Complex Involved in Alternative Splicing Regulation Chen Hung-Hsi, Wang Yu-Chiuan, Fann Ming-Ji. Molecular and Cellular Biology.2006;26(7). CrossRef
- CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1 Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L.. Genes & Development.2010;24(20). CrossRef
- The regulation of glial-specific splicing of Neurexin IV requires HOW and Cdk12 activity Rodrigues F., Thuma L., Klambt C.. Development.2012;139(10). CrossRef
- A New Subfamily of High Molecular Mass CDC2-Related Kinases with PITAI/VRE Motifs Marqués Francois, Moreau Jean-Luc, Peaucellier Gerard, Lozano Jean-Claude, Schatt Philippe, Picard Andre, Callebaut Isabelle, Perret Eric, Genevière Anne-Marie. Biochemical and Biophysical Research Communications.2000;279(3). CrossRef
- CDK13, a New Potential Human Immunodeficiency Virus Type 1 Inhibitory Factor Regulating Viral mRNA Splicing Berro Reem, Pedati Caitlin, Kehn-Hall Kylene, Wu Weilin, Klase Zachary, Even Yasmine, Genevière Anne-Marie, Ammosova Tatiana, Nekhai Sergei, Kashanchi Fatah. Journal of Virology.2008;82(14). CrossRef
- PCTK Proteins: The Forgotten Brain Kinases? Cole Adam R.. Neurosignals.2009;17(4). CrossRef
- Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1 Jiang Mei, Gao Yankun, Yang Tao, Zhu Xueliang, Chen Jiangye. FEBS Letters.2009;583(13). CrossRef
- Wnt Signaling in Mitosis Kaldis Philipp, Pagano Michele. Developmental Cell.2009;17(6). CrossRef
- miR-455 inhibits breast cancer cell proliferation through targeting CDK14 Wang Bing, Zou Aimei, Ma Liqiang, Chen Xiong, Wang Lie, Zeng Ximing, Tan Ting. European Journal of Pharmacology.2017;807. CrossRef
- PFTK1 Promotes Gastric Cancer Progression by Regulating Proliferation, Migration and Invasion Yang Lei, Zhu Jia, Huang Hua, Yang Qichang, Cai Jing, Wang Qiuhong, Zhu Junya, Shao Mengting, Xiao Jinzhang, Cao Jie, Gu Xiaodan, Zhang Shusen, Wang Yingying. PLOS ONE.2015;10(10). CrossRef
- CCRK depletion inhibits glioblastoma cell proliferation in a cilium‐dependent manner Yang Ying, Roine Niina, Mäkelä Tomi P. EMBO reports.2013;14(8). CrossRef
- PCTAIRE-1: Characterization, subcellular distribution, and cell cycle- dependent kinase activity Charrasse S, Carena I, Hagmann J, Woods-Cook K, Ferrari S. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research.1999;10:611-620.
- The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation Besset V, Rhee K, Wolgemuth DJ. Cell Growth Differ.1999;10(3):173-181.
- Multiple Subcellular Localizations of PCTAIRE-1 in Brain Le Bouffant Françoise, Le Minter Pascale, Traiffort Elisabeth, Ruat Martial, Sladeczek Fritz. Molecular and Cellular Neuroscience.2000;16(4). CrossRef
- Cyclin-Dependent Kinase 16/PCTAIRE Kinase 1 Is Activated by Cyclin Y and Is Essential for Spermatogenesis Mikolcevic P., Sigl R., Rauch V., Hess M. W., Pfaller K., Barisic M., Pelliniemi L. J., Boesl M., Geley S.. Molecular and Cellular Biology.2011;32(4). CrossRef
- Two Cyclin-Dependent Kinase Pathways Are Essential for Polarized Trafficking of Presynaptic Components Ou Chan-Yen, Poon Vivian Y., Maeder Celine I., Watanabe Shigeki, Lehrman Emily K., Fu Amy K.Y., Park Mikyoung, Fu Wing-Yu, Jorgensen Erik M., Ip Nancy Y., Shen Kang. Cell.2010;141(5). CrossRef
- CYY-1/Cyclin Y and CDK-5 Differentially Regulate Synapse Elimination and Formation for Rewiring Neural Circuits Park Mikyoung, Watanabe Shigeki, Poon Vivian Yi Nuo, Ou Chan-Yen, Jorgensen Erik M., Shen Kang. Neuron.2011;70(4). CrossRef
- PCTAIRE 2, A Cdc2-Related Serine/Threonine Kinase, is Predominantly Expressed in Terminally Differentiated Neurons Hirose Takashi, Tamaru Teruya, Okumura Nobuaki, Nagai Katsuya, Okada Masato. European Journal of Biochemistry.1997;249(2). CrossRef
- Orphan kinases turn eccentric Mikolcevic Petra, Rainer Johannes, Geley Stephan. Cell Cycle.2012;11(20). CrossRef
- The regulation of tau phosphorylation by PCTAIRE 3: Implications for the pathogenesis of Alzheimer's disease Herskovits A.Z., Davies P.. Neurobiology of Disease.2006;23(2). CrossRef
- PCTAIRE Kinase 3/Cyclin-dependent Kinase 18 Is Activated through Association with Cyclin A and/or Phosphorylation by Protein Kinase A Matsuda Shinya, Kominato Kyohei, Koide-Yoshida Shizuyo, Miyamoto Kenji, Isshiki Kinuka, Tsuji Akihiko, Yuasa Keizo. Journal of Biological Chemistry.2014;289(26). CrossRef
- Human CDK18 promotes replication stress signaling and genome stability Barone Giancarlo, Staples Christopher J., Ganesh Anil, Patterson Karl W., Bryne Dominic P., Myers Katie N., Patil Abhijit A., Eyers Claire E., Maslen Sarah, Skehel J. Mark, Eyers Patrick A., Collis Spencer J.. Nucleic Acids Research.2016;44(18). CrossRef
- The multitalented Mediator complex Carlsten Jonas O.P., Zhu Xuefeng, Gustafsson Claes M.. Trends in Biochemical Sciences.2013;38(11). CrossRef
- A Set of Consensus Mammalian Mediator Subunits Identified by Multidimensional Protein Identification Technology Sato Shigeo, Tomomori-Sato Chieri, Parmely Tari J, Florens Laurence, Zybailov Boris, Swanson Selene K, Banks Charles A.S, Jin Jingji, Cai Yong, Washburn Michael P, Conaway Joan Weliky, Conaway Ronald C. Molecular Cell.2004;14(5). CrossRef
- CDK8 Galbraith Matthew D., Donner Aaron J., Espinosa Joaquín M.. Transcription.2010;1(1). CrossRef
- A Kinase-Independent Role for Cyclin-Dependent Kinase 19 in p53 Response Audetat K. Audrey, Galbraith Matthew D., Odell Aaron T., Lee Thomas, Pandey Ahwan, Espinosa Joaquin M., Dowell Robin D., Taatjes Dylan J.. Molecular and Cellular Biology.2017;37(13). CrossRef
- The Cyclin-Dependent Kinase (CDK) Family Member PNQALRE/CCRK Supports Cell Proliferation but has no Intrinsic CDK-Activating Kinase (CAK) Activity Wohlbold Lara, Larochelle Stephane, Liao Jack C.-F., Livshits Geulah, Singer Juliet, Shokat Kevan M., Fisher Robert P.. Cell Cycle.2006;5(5). CrossRef
- Cell cycle–related kinase is a direct androgen receptor–regulated gene that drives β-catenin/T cell factor–dependent hepatocarcinogenesis Feng Hai, Cheng Alfred S.L., Tsang Daisy P., Li May S., Go Minnie Y., Cheung Yue S., Zhao Gui-jun, Ng Samuel S., Lin Marie C., Yu Jun, Lai Paul B., To Ka F., Sung Joseph J.Y.. Journal of Clinical Investigation.2011;121(8). CrossRef
- Deubiquitinase USP37 Is Activated by CDK2 to Antagonize APCCDH1 and Promote S Phase Entry Huang XiaoDong, Summers Matthew K., Pham Victoria, Lill Jennie R., Liu Jinfeng, Lee Gwanghee, Kirkpatrick Donald S., Jackson Peter K., Fang Guowei, Dixit Vishva M.. Molecular Cell.2011;42(4). CrossRef
- Cyclin-C-dependent cell-cycle entry is required for activation of non-homologous end joining DNA repair in postmitotic neurons Tomashevski A, Webster D R, Grammas P, Gorospe M, Kruman I I. Cell Death & Differentiation.2010;17(7). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2020
Author Details