Exploration of Binding Interaction of Steroidal Drugs with HMG-CoA Reductase for the Treatment of Breast Cancer Patients who suffer with Cardiovascular Problems: In-silico Studies
Download
Abstract
Introduction: HMG-CoA reductase is widely used as a significant target to screen cardiovascular therapeutics molecules because of its involvement in cholesterol synthesis and cell proliferation. Steroidal drugs like anastrozole, exemestane, letrozole have been tested as potential inhibitors of cancer mediated aromatase enzyme and these could be inhibited to HMG-CoA reductase so that the link with cholesterol synthesis and cell proliferation can be utilized to control cancers in.
Objective: To test the binding affinity and binding patterns using computational methods of these drugs and established them as a HMG-CoA reductase inhibitor due to the involvement of this enzyme with cholesterol synthesis and cell proliferation can be utilized to control the cancers.
Materials and methods: Freely available docking tools like AutoDock and web resources like DrugBank and PDB were used to extract data and conduct the study.
Results: The binding interaction of exemestane has shown significant docking interactions which are followed by anastrozole and letrozole. Exemestane has shown binding energy -8.74 kcal/mol, hydrogen bond length: 1.97513 Å, and interacting amino acid was analyzed as A: ASN658:HD21-: UNK1: O6. The positive control statin was used in this study has shown significant binding interaction but it was less than tested exemestane.
Conclusion: Thus, exemestane can potentially inhibit to HMG-CoA reductase enzyme even stronger than the clinically tested drug statin and would be a good choice for cancer patients. Thus, in vitro laboratory experimentation and in vivo research are necessary to put forward therapeutic repositioning of these drugs so they could be established as a broad spectrum potential anticancer drugs including breast cancer especially for the patient with cardiovascular complications.
Introduction
Cancer or tumor among the human population is one of the dangerous diseases worldwide which affected about more than 1 million population and breast cancer is more prevalent in USA among women. The death rate due to cancer is common in the age of 40-59 years [1]. The use of hormonal therapy evolved in 1895 when it was used as a contraceptive drug and pharmacological standardization of steroidal drugs inhibiting the estrogen receptor is the main therapy for the management of breast cancers [2].
Aromatase inhibitors block estrogen synthesis and thus have been tested as well as for therapeuticintervention of breast cancer mediated by estrogen. These inhibit the catalyzing step of the enzyme aromatase complex, which regulates the important step estrogen synthesis [2] and prevent binding of the enzyme with its actual site of the substrate, androgen. Ovaries and many tissues of the body able to synthesize estrogen with the catalyzing power of Aromatase enzymes [3]. The steroidal ones are formestane and exemestane which are structurally similar to natural substrates, testosterone and androstenedione. Expression of aromatase is found in ovaries, extragonadal tissues, brain, fat, liver, endothelium bones and mesenchymal cells of adipose tissue in the breast. Aromatase converts androgens (testosterone and androstenedione) to estrogen and estradiol which is depend on cholesterol synthesis Pathway [4]. Pregnenolone is the precursor of steroid, used in the synthesis of all most all the steroidal hormones. The steroidal hormone synthesis is linked with the cholesterol synthesis which is catalyzed by HMG-CoA reductase, a rate controlling enzyme catalyze the synthesis of cholesterol and mevalonate pathways. The drugs like statins inhibit HMG-CoA reductase enzyme (3-hydroxy-3-methyl-glutaryl-CoA reductase) and applied for HMG-CoA reductase mediated cardiovascular drugs [5]. The drugs like statins have been used to inhibit HMG-CoA reductase enzyme for the management of cardiovascular diseases. Both enzymes are differently linked in a step of cholesterol synthesis. Thus, controlling the cholesterol synthesis could be helpful to manage many types of cancers including breast cancers and ovarian cancers and steroidal drugs are to be tested for search of alternative HMG-CoA reductase inhibitors. Estrogen synthesis is mediated by aromatase which depends on the synthesis of cholesterol. The exemestane inhibits the aromatase enzyme of estrogen synthesis and becomes the choice of drug for the management of breast cancer. Thus, the drugs targeting the aromatase enzyme to inhibit the cholesterol synthesis could be a target to HMG-CoA reductase and hope for emerging inhibitor for the enzyme HMG-CoA reductase. Moreover, It is well reported that the pathogenesis of ovarian cancer is linked with the mevalonate pathway of cholesterol synthesis to potentiate the signaling of the oncogenic pathway [6].Cerivastatin inhibits enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase and contributes significantly to control the spreading of the breast cancer [7].
Chemically it is a 3-hydroxy-3-methylglutaryl coenzyme A reductase and help to convert covert to coenzyme A and mevalonate, a precursor of isoprenoid through the transfer of hydride and utilization two NADPH [8-9]. The researches have described that cholesterol biosynthesis is linked with cancer and cardiovascular disorder. Both experimental and clinical research studied have stated that cancer can be developed due to increased cholesterol synthesis or intake of diet rich in high cholesterol and fat [10-11].
Modification of geranylgeranyl pyrophosphate (GGPP) and/or farnesyl pyrophosphate (FPP) is essential for the function of small GTPases, lamins and other proteins involved in proliferation, survival, invasion, metastasis, inflammation and immune response [12]. Thus, by attenuating mevalonate synthesis, statins not only decrease cholesterol biosynthesis but have pleiotropic effects impinging on multiple cancer pathways [13]. The roles of enzymes aromatase and HMG-CoA reductase enzymes in the development of cancer have been well explored. Simvastatin controls cholesterol synthesis by inhibiting HMG-CoA reductase enzymes and has also been reported for anti-cancer activities. It has potentially induced apoptosis, the cell arrest at G1 and inhibited significantly proliferation of the cell. Moreover, due to inhibition of HMG-CoA reductase enzyme activity which results in cellular stress, DNA damage and inhibition of mTOR and MAPK pathways involved in ovarian cancers [6].
The suppression of ovarian tumor in, orthotropic mouse treated with simvastatin has been reported due to, decreased expression of phosphorylated-p42/44 protein and Ki-67, HMGCR [6].
Thus, Metastatic and invasive properties of cancer cell are linked with HMG-CoA reductase signaling. Moreover, the epigenetic regulation is also affected by HMG-CoA reductase inhibitors [14-15] but extreme cost and length of time in discovery of novel drug and its development are suffering from the low rate of success, the researchers have been accelerated to explore the therapeutic area of existing drugs for new application in oncology.
Materials and Methods
Drug molecule preparation
The three dimensional (3D) configuration of Anastrozole (DB01217), Exemestane (DB00990) and Letrozole (DB01006), and statin (DB00227) like molecules were obtained from DrugBank Database (www.drugbank.ca) [16-18]. It was further minimized by CHARMmforce field [19] minimization of energy procedure through Discovery Studio 2019.
Receptor molecule preparation
The well-known protein structure database Protein Data Bank(PDB) governed by Research Collaborator of Structural Bioinformatics (RCSB) (www.rcsb.org) used to download the 3D structure of HMG-Co A reductase (PDB ID: 1DQ9). The structure was selected based on available experimental data like X-ray diffraction method used to develop the 3D structure with the obtained resolution of 2.80 Å, R-value Free was 0.247 and R-value work was 0.207. The water molecules and HETATM were removed from the PDB ID: 1DQ9. file and CHARMm force field applied for energy minimization [19] using Discovery Studio 2019 [19-20].
In silico interaction analysis
MGL tool Autodock has been used to analyze the binding interaction between drug compounds and HMG-CoA reductase. The analysis of interaction was carried out through Lamarckian genetic algorithm (LGA) Molecular docking methods and then probing the best conformation of enzyme and drug complex on the calculation is done and obtained binding energy (∆G).
∆G binding = ∆Ggauss + ∆Grepulsion + ∆Ghbond +∆Ghydrophobic + ∆Gtors
Here, ∆G gauss is dispersion of two gaussian functions, ∆G repulsion is square of the distance if closer than a threshold value, ∆Ghbond & ∆Ghydrophobic are ramp function, also used for interactions with metal ions and ramp function respectively while ∆Gtors is relative to the no. of rotatable bonds [21-22].
Further water molecules were removed from the 3D structure of enzyme (PDB ID: 1DQ9) before docking and hydrogen atoms and finally salvation parameters and charge (Kollman united) were added to the PDB ID: 1DQ9. Drug compounds with Gasteiger charge was used to conduct the study. To cover highest part of drug compounds and PDB ID: 1DQ9, a Grid box set was selected and set to 60×60×60° values at all three axis of grid point. A 0.375 Å of space was selected as default grid points. The docking analysis for (PDB ID: 1DQ9)-drug compounds was performed through Lamarckian Genetic Algorithm (LGA) [23-24].
The default LGA parameters like population size (ga_pop_size) 150, energy evaluations (ga_num_ generation) 2500000, mutation rate 27000, crossover rate 0.02 and 0.8 and step size 0.2 Å were set and 10 runs of LGA set to complete the study. After successful execution of docking steps obtained conformations of PDB ID: 1DQ9 and drug compounds complexes were analyzed for the interactions and binding energy using Discovery Studio 2019 molecular visualization software.
Results
Thus, in this study, Insight structural recognition and binding pattern analysis of HMG-CoA reductase with drugs like exemestane, anastrozole, and letrozole have been conducted to explore the therapeutic use of these drugs to control the ovarian and breast cancers. Recently published reports have shown that exploration of therapeutics uses of established drugs have been accelerated. Similarly, explorations of therapeutics use of these drugs conducted in this study, have shown the significant binding affinities with enzyme HMG-CoA. Among the studied drugs the exemestane has showed the maximum negative binding energy -8.74 kcal/mol Figure 1b; Table 1. The enzyme ligand interaction was facilitated by hydrogen bond observed at A:ASN658:HD21-:UNK1:O6H position and observed bond distance was 1.97513 Å. The hydrophobic binding interaction also observed with the involved Met 655, Gly656, Met657, Asn658, Met659, Tyr761, Gln766, Asp767, Gln770, Glu801, Ile802, Gly803, Thr804, Val l805, Gly80, Gly807, Gly808, Thr809 amino acids. The observed values for inhibition constant is 395.14 nM.
| S.No | Hydrogen Bond | Hydrogen Bond Distance (Å) | Binding Energy | Inhibition Constant | Residues involved in Hydrophobic interaction |
| 1. Anastrozole | A:LYS691:HZ3 - :UNK0:N2 | 2.33883 | -7.84 kcal/mol | 13.93uM | Cys526, Met655, Met657, Asp690, |
| :UNK0:N3 - A:GLN766:O | 2.90861 | Lys691, Tyr761, Gly765, Gln766, | |||
| :UNK0:H6 - A:MET655:SD | 2.68592 | Asp767, Gln770, Glu801, Ile802, | |||
| :UNK0:H16 - A:GLN766:OE1 | 1.9906 | Gly803, Gly806, Gly807, | |||
| :UNK0:H17 - A:GLU801:O | 2.2311 | Gly808 | |||
| 2. Exemestane | A:ASN658:HD21 - :UNK1:O6 | 1.97513 | -8.74 kcal/mol | 395.14 nM | Met655, Gly656, Met657, Asn658, |
| Met659, Tyr761, Gln766, Asp767, | |||||
| Gln770, Glu801, Ile802, Gly803, | |||||
| Thr804, Val805, Gly80, Gly807, | |||||
| Gly808, Thr809 | |||||
| 3. Let | A:MET655:HN - :UNK0:N2 | 2.82966 | -6.66 kcal/mol | 108.27 uM | Cys526, Arg590, Met655, Met657, |
| :UNK0:H1 - A:GLY765:O | 2.10095 | Asp690, Lys691, Gly765, Gln766, | |||
| :UNK0:H2 - A:ASP690:OD2 | 1.72832 | Asp767, Gln770, Gly803, Val805, | |||
| :UNK0:C14 - A:GLN766:OE1 | 3.71849 | Gly807, Gly808, Thr809 | |||
| :UNK0:C14 - A:GLY803:O | 3.65332 | ||||
| :UNK0:C14 - :THR809:OG1 | 3.68008 | ||||
| Control | |||||
| Statin | A:ASN658:HD21 - :UNK0:O3 | 2.23592 | -7.16 kcal/mol | 5.63 uM | Cys526, Met655, Gly656, Met657, |
| :UNK0:H2 - A:GLY803:O | 3.00617 | Asn658, Gly765, Gln766, Asp767, | |||
| :UNK0:H2 - A:THR809:OG1 | 1.87243 | Gln770, Glu801, Ile802, Gly803, | |||
| A:ILE802:CA - :UNK0:O4 | 3.38298 | Thr804, Val805, Gly806, Gly807, | |||
| A:GLY807:CA - :UNK0:O | 3.66334 | Gly808, Thr809 | |||
The docking features signified the HMG-CoA reductase inhibitory potential of studies letrozole, anastrozole, exemestane are shown in Figure 2b-3b; Table 1.
The anastrozole has also been shown significant binding interaction with HMG-CoA reductase and binding energy of the enzyme ligand was -7.84 kcal/mol (Figure 1; Table 1) but hydrogen bonding facilitated was due to involvement of amino acids which are as A:LYS691:HZ3-:UNK0:N2:UNK0:N3- A:GLN766:O:UNK0:H6-A:MET655:SD:UNK0:H16 A:GLN766:OE1:UNK0:H17,:GLU801:O amino acids and distance for hydrogen bond were 2.33883, 2.90861, 2.68592, 1.9906, 2.2311 Å, respectively.
Figure 1: 3D Graphics Showing (a) Ana (b) exe and (c) Let (shown in green stick pattern) Interaction with HMG receptor Amino acid Residues (shown by surrounded stick pattern) Involved in Hydrophobic Interaction. Blue dotted lines showing hydrogen bonds and length in Angstrom. Graphics generated by Discovery Studio Visualizer. ana, anastrozole; exe, exemestane and let, letrozole..
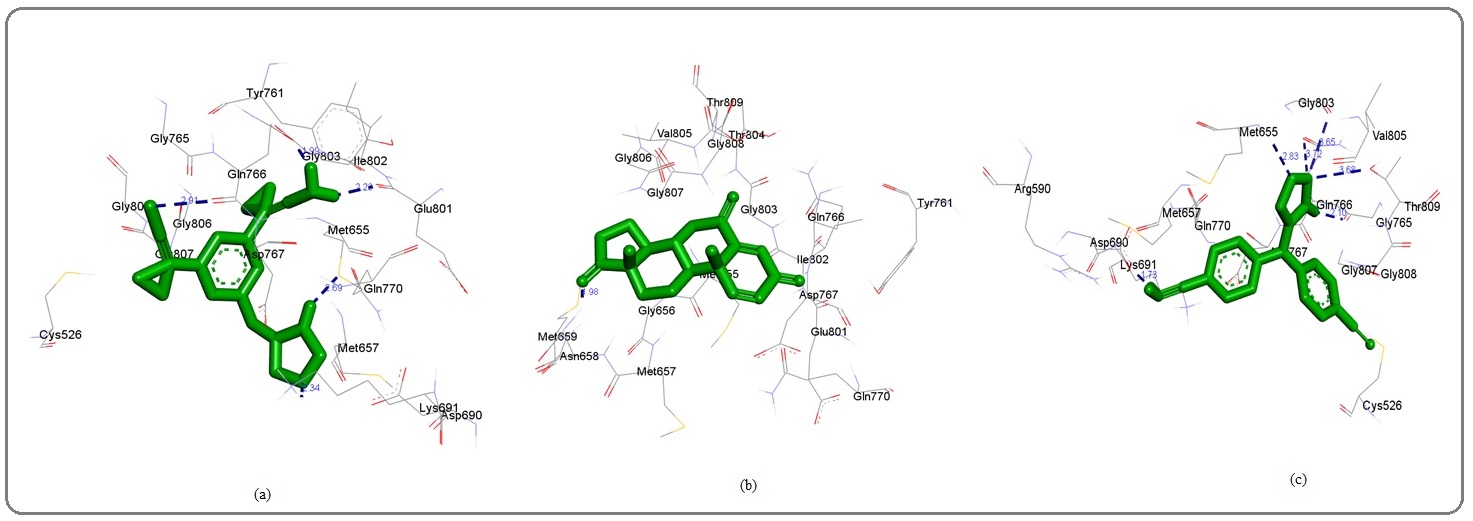
The inhibition constant was 13.93uM and observed amino residues faceplate the hydrophobic interaction were Cys526, Met655, Met657, Asp690, Lys691, Tyr761, Gly765, Gln766, Asp767, Gln770, Glu801, Ile802, Gly803, Gly806, Gly807, Gly808 (Figure 2; (Figure 1a- Figure 2a and 3a; Table1). The letrozole has shown minimum binding interaction with the enzyme and analyzed binding energy was -6.66 kcal/mol (Table 1).
Figure 2: 2D Graphics Showing (a) Ana (b) Exe and (c) Let (shown in green stick pattern in the middle) Interaction with HMG Receptor Amino acid Residues (shown by surrounded round shaped pattern) Involved in Different Types of Molecular Interaction. Graphics Generated by Discovery Studio Visualizer. ana, anastrozole; exe, exemestane and let, letrozole..
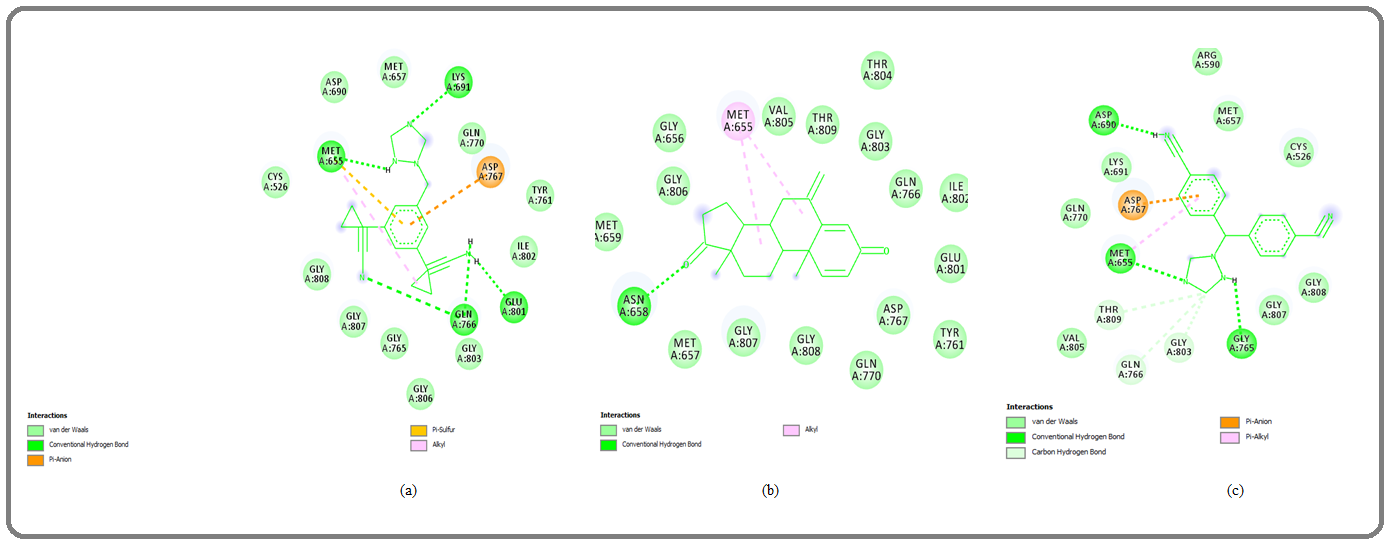
The observed hydrogen bond forming residues are A:MET655:HN-:UNK0:N2 with a distance 2.82966 Å, UNK0:H1-A:GLY765:O (2.10095 Å), UNK0:H2- A: ASP690:OD2 (1.72832 Å), UNK0:C14- A:GLN766:OE1 (3.71849 Å),UNK0:C14-A:GLY803:O (3.65332 Å),: UNK0:C14-A:THR809:OG1 and 3.68008 Å hydrophobic interaction the involved amino acids are Cys526, Arg590, Met655, Met657, Asp690, Lys691, Gly765, Gln766, Asp767, Gln770, Gly803, Val805, Gly807, Gly808, Thr809 (Figure 1c and Figure 3c;).
Figure 3: 3D Graphics Showing (a) Ana (b) Exe and (c) Let (shown in green stick pattern) Interaction with HMG Receptor Pocket. Blue Dotted Lines Showing Hydrogen Bonds and Length in Angstrom. Graphics Generated by Discovery Studio Visualizer. ana, anastrozole; exe, exemestane and let, letrozole..
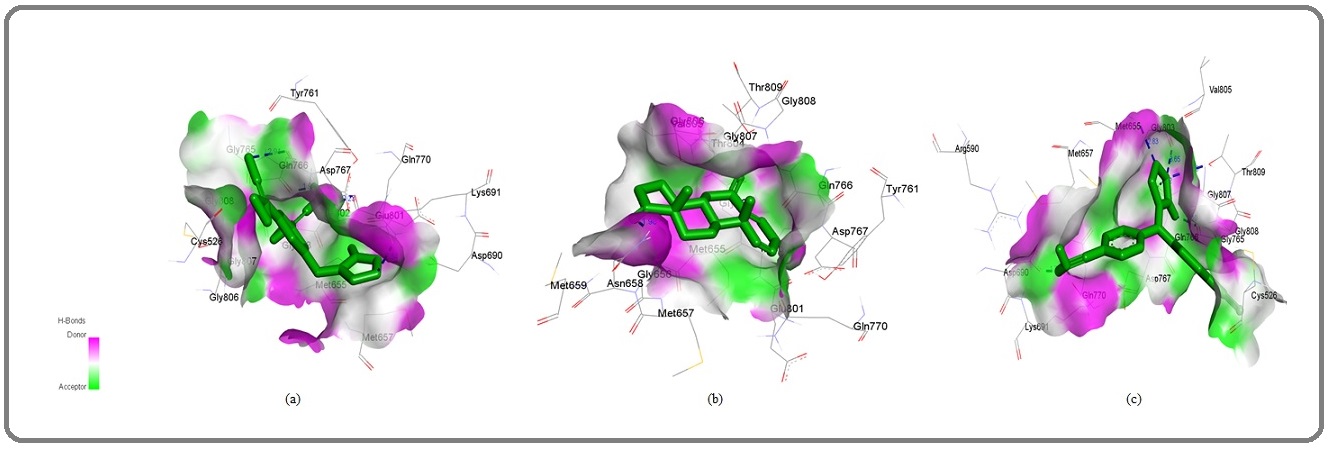
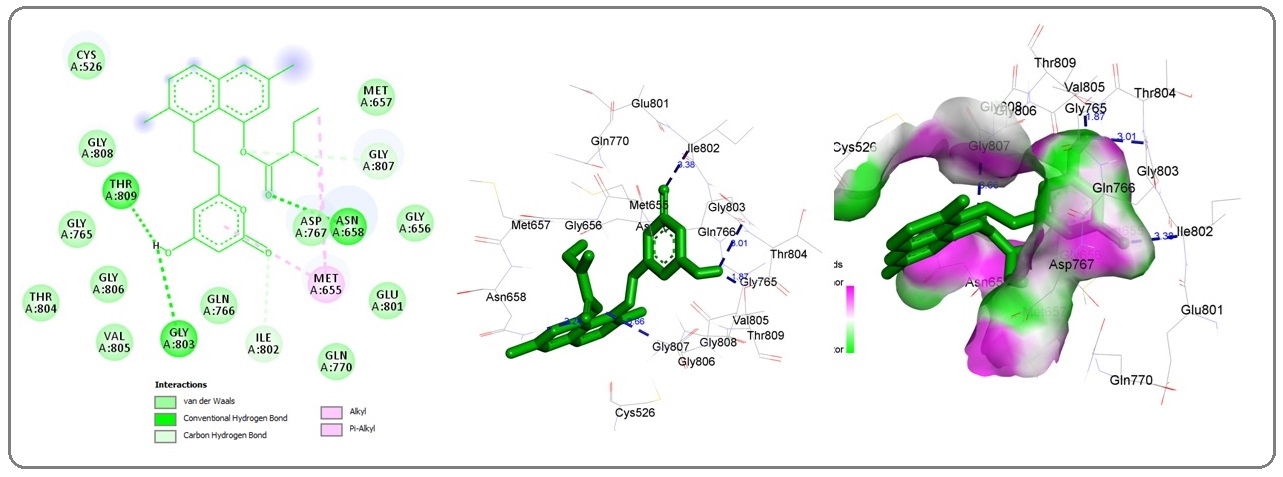
The observed binding energy of statin with HMG-CoA reductase was observed to be-7.16 kcal/mol, hydrogen bond length 2.23592 A0,and hydrophobic interaction was facilitated by A:ASN658:HD21 - :UNK0:O3, Cys526, Met655, Gly656, Met657, Asn658, Gly765, Gln766, Asp767, Gln770, Glu801, Ile802, Gly803, Thr804, Val805, Gly806, Gly807, Gly808,Thr809 amino acids (Figure 4a-c).
Figure 4: 3D Graphics Showing Statin Interaction (shown in green stick pattern) Interaction with HMG Receptor Amino acid Residues (shown by surrounded stick pattern) Involved in Hydrophobic Interaction. Blue Dotted Lines Showing Hydrogen Bonds and Length in Angstrom. c, Statin Interaction with HMG-COA Reductase Pocket, Graphics Generated by Discovery Studio Visualizer..
Discussion
The drugs targeting the aromatase enzyme to inhibit the cholesterol synthesis could be target to HMG-CoA reductase to control the cholesterol synthesis and could be a hope for these as emerging inhibitor of HMG- CoA reductase. Moreover, the mevalonate pathway linked with cholesterol, is thought to be a potential oncogenic pathway in the pathogenesis of ovarian cancer [6]. Molecular mechanism of the anti-cancer activity of cerivastatin, an inhibitor of HMG-CoA reductase, on aggressive human breast cancer cells [7]. HMG- CoA reductase, a 3-hydroxy-3-methyl-glutaryl- CoA reductase is involved in synthesis of cholesterol. Cholesterol linked with mevalonate pathway which is reported to be a key oncogenic series of the reaction of the progression of ovarian cancer [7]. The use of statins in cancer was described in the early 1990s. The pro-apoptotic and anti-proliferative effects of statin have been well explored in many animal models and different types of cell lines. Many anti-tumorigenic drugs have been widely used as HMG-CoA reductase inhibitors to control cholesterol synthesis. HMG-CoA reductase involved in cellular proliferation, cell cycle regulation. The drugs like simvastatin inhibiting HMG-CoA reductase have been well reported to control cell proliferation, induced apoptosis and DNA damage as well as cell arrest at G1 phase including inhibition of the mTOR and MAPK pathways to control ovarian cancers. The inhibition of this enzyme also decreases the level of cholesterol, low-density lipoprotein (LDL) and suppression of atherosclerotic plaque synthesis [25]. It has been earlier reported that many steroidal drugs inhibit the cholesterol synthesis [10]. The non-steroidal drugs like exemestane, anastrozole, and letrozole have replaced tamoxifen and become the choice of treatment for the estrogen receptor of the breast cancer [26].
These drugs already have also been used as a choice of breast cancer therapeutics. The interaction analyses from this study explore the therapeutic area of these drugs. The docking study represented a significant binding of these drugs with HMG-CoA reductase and could be used as a choice of drugs for the treatment of ovarian cancers, breast cancer and cardiovascular diseases. The results of this study indicated that these exemestanes has maximum binding energy which is followed by anastrozole and letrozole and could be used as inhibitors to control cholesterol mediated cancers. Recently, many natural compounds and their in silico inhibitory binding interaction with HMG-CoA reductase has been described [27-28]. Moreover, small-molecules like rottlerin and Statin were reported as HMG-CoA reductase inhibitors to prevent metastasis and progression of colon cancer via MACC1 [29]. The concentration and time-dependent anticancer effect of lovastatin, mevastatin, and rottlerin were reported due to suppression of MACC1 mRNA and protein expression [29]. Moreover, it also was described as the anticancer potentiality of HMG-CoA inhibitors [30]. Thus, based on above finding it is concluded that these drugs could be used as HMG-CoA reductase inhibitors and they could be a choice of drugs not only in the treatment of breast cancers but also in the management of colon cancer, ovarian cancers, brain tumors, pancreatic cancer, castration-resistant prostate cancer, lung cancer like those cancers where cholesterol play a significant role in the progression of cancers. Moreover, they also have been reported to increase the synthesis of proapoptotic protein Bax but decrease antiapoptotic protein Bcl-2 synthesis [31-33]. Thus, these drugs in a combination of meclofenamic acid and simvastatin could be a significant strategy for the treatment of cancers including castration-resistant prostate cancer.
In conclusion, this is the first study exploring the new target of drugs for the treatment of cancers and these drugs could be an alternative choice to inhibit HMG-CoA reductase and their use in cancer patients especially to the patient with cardio vascular complication. HMG-CoA reductase inhibitors play a significant role in the management of cardiac diseases. Statins are some of the most widely used medications in the world with significant input for primary and secondary prevention of cardiovascular diseases. The anticancer properties of these drugs have also been suggested and are attracting increasing interest among the biomedical society. Although, these drugs are safely used to manage these disease but due to short elimination, fast achieving concentration and low systemic bioavailability of these molecules make an need to search new cheap and potential anticancer molecules. On the whole, this in silico study suggest that exemestane, anastrozole and letrozole could be used as HMG-CoA inhibitors and might be promising potential cancer therapeutic, given a hope for fast clinical translation.
Thus, these studies molecules are well tested molecules and their in vitro-in vivo exploration will be helpful for the management of many types of cancers. Further investigations are needed for an advanced clarification of the antitumor potential of statins (including particular cancer type (s), dosage and type of statin, treatment schedule and biomarkers to predict patient’s sensitivity to this class of medications, etc.
Acknowledgements
The author is thankful to the Health Information Technology Department, Faculty of Applied Studies and Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under grant No. (D1441-302-130). The authors, therefore, gratefully acknowledge the DSR technical and financial support.
Conflict of Intrest
The author declares that there is no conflict of interest.
References
- Global Cancer in Women: Burden and Trends Torre Lindsey A., Islami Farhad, Siegel Rebecca L., Ward Elizabeth M., Jemal Ahmedin. Cancer Epidemiology Biomarkers & Prevention.2017;26(4). CrossRef
- Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer Jordan V. Craig, Brodie Angela M.H.. Steroids.2007;72(1). CrossRef
- Tissue physiology and pathology of aromatase Stocco Carlos. Steroids.2012;77(1-2). CrossRef
- Aromatase expression and regulation in breast and endometrial cancer Zhao Hong, Zhou Ling, Shangguan Anna Junjie, Bulun Serdar E. Journal of Molecular Endocrinology.2016;57(1). CrossRef
- Controlling Cholesterol Synthesis beyond 3-Hydroxy-3-methylglutaryl-CoA Reductase (HMGCR) Sharpe Laura J., Brown Andrew J.. Journal of Biological Chemistry.2013;288(26). CrossRef
- The HMG-CoA reductase inhibitor, simvastatin, exhibits anti-metastatic and anti-tumorigenic effects in ovarian cancer Stine Jessica E., Guo Hui, Sheng Xiugui, Han Xiaoyun, Schointuch Monica N., Gilliam Timothy P., Gehrig Paola A., Zhou Chunxiao, Bae-Jump Victoria L.. Oncotarget.2015;7(1). CrossRef
- Cholesterol and Its Metabolites in Tumor Growth: Therapeutic Potential of Statins in Cancer Treatment Chimento Adele, Casaburi Ivan, Avena Paola, Trotta Francesca, De Luca Arianna, Rago Vittoria, Pezzi Vincenzo, Sirianni Rosa. Frontiers in Endocrinology.2019;9. CrossRef
- The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases Friesen J, Rodwell V. Genome biology.2004;5(11):248.
- QM/MM study of the mechanism of reduction of 3-hydroxy-3-methylglutaryl coenzyme A catalyzed by human HMG-CoA reductase Eduardo F. Oliveira , NMFSAC , Maria J. Ramos , Pedro A. Fernandes . Catal. Sci.2016.
- The role of cholesterol metabolism and cholesterol transport in carcinogenesis: a review of scientific findings, relevant to future cancer therapeutics Cruz Pedro M. R., Mo Huanbiao, McConathy Walter J., Sabnis Nirupama, Lacko Andras G.. Frontiers in Pharmacology.2013;4. CrossRef
- Cholesterol Lowering in Cancer Prevention and Therapy CE HCaF . Cholesterol Lowering Therapies and Drugs .2016.
- Inhibition of farnesyl pyrophosphate (FPP) and/or geranylgeranyl pyrophosphate (GGPP) biosynthesis and its implication in the treatment of cancers Waller Daniel D., Park Jaeok, Tsantrizos Youla S.. Critical Reviews in Biochemistry and Molecular Biology.2019;54(1). CrossRef
- HMG-CoA Reductase Inhibition Delays DNA Repair and Promotes Senescence After Tumor Irradiation Efimova Elena V., Ricco Natalia, Labay Edwardine, Mauceri Helena J., Flor Amy C., Ramamurthy Aishwarya, Sutton Harold G., Weichselbaum Ralph R., Kron Stephen J.. Molecular Cancer Therapeutics.2017;17(2). CrossRef
- Pleiotropic and Adverse Effects of Statins—Do Epigenetics Play a Role? Allen Stephanie C., Mamotte Cyril D.S.. Journal of Pharmacology and Experimental Therapeutics.2017;362(2). CrossRef
- Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells Karlic Heidrun, Thaler Roman, Gerner Christopher, Grunt Thomas, Proestling Katharina, Haider Florian, Varga Franz. Cancer Genetics.2015;208(5). CrossRef
- DrugBank 5.0: a major update to the DrugBank database for 2018 Wishart David S, Feunang Yannick D, Guo An C, Lo Elvis J, Marcu Ana, Grant Jason R, Sajed Tanvir, Johnson Daniel, Li Carin, Sayeeda Zinat, Assempour Nazanin, Iynkkaran Ithayavani, Liu Yifeng, Maciejewski Adam, Gale Nicola, Wilson Alex, Chin Lucy, Cummings Ryan, Le Diana, Pon Allison, Knox Craig, Wilson Michael. Nucleic Acids Research.2017;46(D1). CrossRef
- Dassault Systèmes BIOVIA (2019) Discovery Studio Visualizer, version 19.1.0.18287, San Diego: Dassault Systèmes .
- The Protein Data Bank Berman H. M.. Nucleic Acids Research.2000;28(1). CrossRef
- CHARMM: The biomolecular simulation program Brooks B. R., Brooks C. L., Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M.. Journal of Computational Chemistry.2009;30(10). CrossRef
- Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis Istvan Eva S., Palnitkar Maya, Buchanan Susan K., Deisenhofer Johann. The EMBO Journal.2000;19(5). CrossRef
- Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function Morris GM G, Halliday R, Huey R, Hart W, Belew R, et al . Journal of computational chemistry.1998;19:1639-1662.
- AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility Morris Garrett M., Huey Ruth, Lindstrom William, Sanner Michel F., Belew Richard K., Goodsell David S., Olson Arthur J.. Journal of Computational Chemistry.2009;30(16). CrossRef
- Automated docking of flexible ligands: Applications of autodock Goodsell David S., Morris Garrett M., Olson Arthur J.. Journal of Molecular Recognition.1996;9(1). CrossRef
- An improved LGA for protein ligand docking prediction Tsai CW C, Yang C. Proc IEEE Congr Evol Comput.2012;:1-6.
- The Role of Statins in Cancer Therapy Hindler Katja, Cleeland Charles S., Rivera Edgardo, Collard Charles D.. The Oncologist.2006;11(3). CrossRef
- The what, why and how of aromatase inhibitors: hormonal agents for treatment and prevention of breast cancer Fabian C. J.. International Journal of Clinical Practice.2007;61(12). CrossRef
- Natural Inhibitors of HMG-CoA Reductase-An Insilico Approach Through Molecular Docking and Simulation Studies Suganya Subramanian, Nandagopal Balaji, Anbarasu Anand. Journal of Cellular Biochemistry.2016;118(1). CrossRef
- Eight new triterpenoids with inhibitory activity against HMG-CoA reductase from the medical mushroom Ganoderma leucocontextum collected in Tibetan plateau Zhang Jinjin, Ma Ke, Han Junjie, Wang Kai, Chen Hongyu, Bao Li, Liu Li, Xiong Weiping, Zhang Yaodong, Huang Ying, Liu Hongwei. Fitoterapia.2018;130. CrossRef
- Statin and rottlerin small-molecule inhibitors restrict colon cancer progression and metastasis via MACC1 Juneja Manisha, Kobelt Dennis, Walther Wolfgang, Voss Cynthia, Smith Janice, Specker Edgar, Neuenschwander Martin, Gohlke Björn-Oliver, Dahlmann Mathias, Radetzki Silke, Preissner Robert, von Kries Jens Peter, Schlag Peter Michael, Stein Ulrike. PLOS Biology.2017;15(6). CrossRef
- HMG-CoA reductase inhibitors (statins) as anticancer drugs (review) Fritz G. International journal of oncology.2005;27(5):1401-1409.
- HMG-CoA Reductase Inhibition Delays DNA Repair and Promotes Senescence After Tumor Irradiation Efimova Elena V., Ricco Natalia, Labay Edwardine, Mauceri Helena J., Flor Amy C., Ramamurthy Aishwarya, Sutton Harold G., Weichselbaum Ralph R., Kron Stephen J.. Molecular Cancer Therapeutics.2017;17(2). CrossRef
- Molecular mechanism of the anti-cancer activity of cerivastatin, an inhibitor of HMG-CoA reductase, on aggressive human breast cancer cells Denoyelle Christophe, Albanese Patricia, Uzan Georges, Hong Li, Vannier Jean-Pierre, Soria Jeannette, Soria Claudine. Cellular Signalling.2003;15(3). CrossRef
- Simvastatin inhibits cancer cell growth by inducing apoptosis correlated to activation of Bax and down-regulation of BCL-2 gene expression SPAMPANATO CARMINE, DE MARIA SALVATORE, SARNATARO MADDALENA, GIORDANO ELISABETTA, ZANFARDINO MARIO, BAIANO SALVATORE, CARTENÌ MARIDELA, MORELLI FRANCO. International Journal of Oncology.2011;40(4). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2020
Author Details