Molecular Genetics of Esophageal Cancer : Indian Perspective
Download
Abstract
Background: RIZ1 is one of the tumor-suppressor genes that is silenced in many human cancers. Change in RIZ1 expression has not been reported in ESCC patients. Therefore, the aim of this study was to investigate the role of RIZ1 in ESCC in the Indian population.
Methods: Twelve esophageal squamous-cell carcinoma (ESCC) patients in stage IV and 12 healthy individuals were used in this study. Tissue sampling was taken from individuals and total RNA was isolated and then cDNA was synthesized using PCR. RIZ1 primers were then designed, and RIZ1 expression was quantified by qRT-PCR. Mapping of CpG islands in RIZ1 promoter was performed using bioinformatics tools. The promoter methylation status of this gene was studied using u methylation-specific PCR (MSP). T-student test was used to analyze the data.
Results: Decreased RIZ1 expression was observed in ESCC compared with healthy controls. The results showed a relatively higher density of CpG dinucleotides in the RIZ1 promoter. No significant difference in promoter methylation was observed in blood and tissue samples.
Conclusion: The study showed a significant down-regulation of RIZ-1 gene in the blood and tissue samples of ESCC patients that did not related to the altered promoter methylation.
Introduction
Over expression of Cyclin B1 in ESCC has been reported to induce tumor cell invasive growth and metastasis [1]. PTEN may play a role in carcinogenesis and progression of ESCC in a high incidence area of northern China [2]. Altered Ras function can contribute to human esophageal SCC development [3]. EMP3 may be a tumor suppressor gene at the last step of esophageal carcinogenesis [4]. Down-regulation of RUNX3 may play a role in disease progression of ESCC [5].
The genetic mechanisms that cause Esophageal squamous cell carcinoma (ESCC) have been attributed to changes in genes encoding tumor-suppressing proteins including P53, disruption of the cell division cycle in the G1/S phase, and deregulation of cell division control and increased oncogene expression. Other genes involved in the development of ESCC include Cyclin B1, which is involved in the metastatic state of the disease [1], PTEN, which plays an important role in the development of this cancer [2], and other genes such as ALCAM, BPA-2, MMP7, MMP13, and MMP9 are also noted. Another ESCC mechanism is CpG island hypermethylation in the gene encoding E-cadherin, which has been associated with poor prognosis in these patients. Down-regulation of the TGM3 gene is another event that has occurred in the ESCC and has been reported to play an important role in this process. Other important genes in the ESCC development process include altered Ras function [3], overexpression of Aurora-A [6], Fra-1, Id-1, and CDC25B.
The role of the VEGF-C gene in ESCC progression through angiogenesis has been reported COX-2 is another gene involved in regulating the proliferation and apoptosis of ESCC tumor cells and has been shown to overexpressed in this disease. Down-regulation of the EMP3 [7] and RUNX3 [5] genes, which are tumor-suppressing genes, has been reported in the ESCC as well. Overexpression of LDC25B and LAMC2 genes was reported in the Chinese population at high risk for ESC. The inhibition of FHT gene expression in alcohol-induced esophageal carcinogenesis and multicentric carcinogenesis has been reported. One of the oncogenes that has been considered for its role in ESCC progression and metastasis is the Stefin A, which is hoped to be used as a therapeutic target for ESCC in the future.
In ESCC, changes in the expression of Arachidonic acid metabolism pathway genes play an important role in its development. Therefore, the expression of genes involved in this pathway can be used as a biomarker in esophagus carcinogenesis, which can be referred to DDH [8], Surviv [9], OPTN [10], MAP3K3, AKAP13, ZnT7 and TG2 [11] over expression.
Comparative Genome Hybridization (CGH) and loss of heterozygosity (LOH) are among the methods of analysis of the whole genome or specific gene, which in the ESCC indicates genetic complexity and many changes in the copy number. Recent studies have shown an increase in the copy number of oncogenes such as EGFR, MYC, FGFR, CCND1, FGF4 / 3, and EMS1 and a decrease in the copy number in the FHIT, APC, RB1 and CDKN2A genes. This has been associated with poor prognosis in the ESCC, however, this relationship has been weak.
B. Genetic alterations reported in ESCC of High-risk region in India
In the Assam region of India, down-regulation of genes involved in ribosome structure formation and genes in cation- transporter activity, GPCR and MAPK was observed in non-familial ESCC cases [12]. However, this clustering in the high-risk area of India seems to be more due to common environmental factors rather than genetic factors.
Copy number alteration study revealed that candidate genes in the potential signalling pathways (focal adhesion, tight junction, the MAPK signalling pathway and the neuroactive ligand receptor interaction pathway) might be used as molecular biomarkers in the early detection of ESCC in high-risk area of India in association with tobacco and betel quid use.
Studies have shown that in some genes involved in signalling pathways, changes in the copy number occur, which has the potential to be used as a biomarker in the early detection of ESCC in high-risk areas of India [13]. Increased copy number of BRCA2 [14] and EPHX1 [15] genes has been reported to play a role in ESCC in high-risk areas of India.
One of the genes involved in the development of ESCC is the TNFα gene. The TNFα-308GA variant increases ESCC risk in female patients with lymph node interference, while the other variant of this NFKBIA-826CT + TT gene reduces the risk of ESCC in women. A reduction in the risk of this cancer has also been reported in haplotypes such as NFKBIA -826C> T, 3’UTRA> G polymorphisms, C-826G3’UTR and T-826A3’UTR. Some polymorphisms, such as TNFα-308 and NFKBIA (-826C> T and 3’UTRA> G), have been shown to increase hypersensitivity and have not been associated with patient prognosis.
One of the genes that has recently been found to inhibit tumor growth is the Esophageal cancer-related gene 1 (ECRG1). Some polymorphisms in this gene, such as ECRG1 Arg 290 Gln, increase sensitivity to ESCC [16]. Some variants of CASP8 gene such as CASP8 IVS12-19 G> A are involved in increasing the risk of ESCC and survival in ESCC patients [16]. GSTP1val/ val and CYP2E1c1c2 polymorphisms have increased the risk of ESCC, but another variant of this GSTM3AB gene has been associated with a reduced risk of ESCC [17]. The haplotypes C (481) A (590) G (857) and T (481) A (590) G (857) NAT2 gene [17] and COX-2 polymorphism -765G> C [18] has been shown to increase sensitivity to ESCC.
Esophageal cancer is an aggressive malignancy with a poor prognosis. EC forms have varied depending on the geographical area. For example, ESCC is the most common form of the disease in northern China and South Africa, while in Europe and the United States adenocarcinoma form is more common [19]. Mutations have been reported in genes such as p53 and p16, amplification of D-cyclin, Myc, EGFR, and deletion genes [20-21]. The aim of the present research was to study the expression profile of Retinoblastoma protein-interacting zinc finger gene (RIZ) in patients with EC.
Reason for choosing Retinoblastoma Protein-interacting zinc finger gene (RIZ1)
Retinoblastoma Protein-interacting zinc finger gene (RIZ) maps to human chromosome 1 (1p36). RIZ1 is considered as a tumour suppressor gene (TSG) because RIZ1 can induce G2-M arrest and apoptosis. The expression of RIZ1 is frequently silenced in many human malignant tumours and reduced RIZ1 expression increased the chance of metastasis. With this background and moreover as there were no studied done on RIZ1 in the Indian population we chose to observe its role in EC patients of Indian origin.
RIZ is a 220-kDa protein, containing eight zinc finger motifs and maps to the distal short arm of human chromosome 1 (1p36), a region has been shown to harbour several tumor suppressor genes implicated in human malignancies. The product of RIZ gene is two protein isoforms RIZ1 and RIZ2, with different lengths [22]. The former has a positive second regulator and acts as a tumor suppressor in many human cancers because it blocks the cell division cycle and induces apoptosis. RIZ1-deficient mice develop an unusual spectrum of tumours and in a genetic background compromised for p53, they suffer accelerated tumor formation and mortality. A strong positive correlation has been reported between down-regulation RIZ1 gene and increased risk of metastasis. Although role of RIZ1 as a tumour suppressor has been established in breast cancer, gastric cancer, it’s role in human EC has been recently explored. In general, RIZ-1 expression is frequently abolished in many human malignant tumours. The distal short arm of chromosome 1 is seen in a human neoplasm Besides, Loss of RIZ-1 gene function in a variety of cancers including gastric, esophagus and breast cancers has been attributed to loss of heterozygosity (LOH), promoter hypermethylation, and frameshift mutations. Given this brief background, we put forward following objectives.Although role of RIZ1 as a tumour suppressor has been established in breast cancer. The aim of this study was to 1) compare the expression analysis of RIZ1 gene between EC patients and healthy controls, 2) to compare the expression analysis of RIZ1 gene in peripheral blood lymphocytes and affected tissue samples in EC patients, and 3). To evaluate the methylation status of CpG dinucleotides at RIZ1 promoter.
Materials and Methods
A. Recruitment of study Participants
The present study was conducted in two phases. In the first phase a reduced RIZ-1 gene expression was documented in the patient’s (EC Stage IV) blood samples as compared to the normal subjects. For this, 12 oesophageal cancer (EC) patients and 12 normal individuals without cancer were recruited from Cancer Care Welfare Home (CCWHRI), Kolkata. Blood samples were collected with appropriate informed consent. All had stage IV cancer. Controls who were never diagnosed with cancer were recruited from Fortis Hospital OPD from men and women.
In the second phase, RIZ-1 expression was compared in paired samples i.e. between the PBMC fraction of blood and biopsy proven cancer tissue of the same patient. For this, 12 esophageal cancer (EC) patients were selected from NCRI, Kolkata. While performing the biopsy for a part of the diagnosis tissue samples were collected. Blood samples were also collected from the same individuals.
B. RNA and DNA isolation
Total RNA from tissue (>50 mg) was extracted using TRI reagent (Sigma Aldrich, St. Louis, MO) using standard protocol. The quantity and quality of extracted RNA was examined spectrophotometrically. DNA isolation was done from 100mg of tissue samples stored in RNA later solution at -800C degree using QIAamp DNeasy Mini Kit (QIAGEN)and eluted DNA was dissolved in 200µl of AE buffer and stored at 40C for further use. Genomic DNA was also isolated from 100µl of blood following instructions of QIAamp DNeasy Mini Kit (QIAGEN) and eluted DNA was dissolved in 200µl of AE buffer and stored at 40C until further use.
C. Quantitative Gene Expression assay
Reverse transcription of 1μg of RNA sample isolated from blood or tissue was random hexamers and High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems Inc) as the template for qPCR reactions.
The primers used for expression analysis of RIZ1 are: forward 5’TCTGCTGTTGACAAGACCC3’ and reverse 5’GCATCAATGCACATCCATC3’ yielding a product size of 167bp. The PCR comprised of 40 cycles of denaturation at temperature of 940C for 30s, annealing at temperature of 550C for 30s and elongation at 720C for 30s.
Mapping of CpG islands in RIZ1 promoter: We selected the sequence of 1200bp of RIZ1 promoter, 200bp upstream of the transcriptional start site (TSS) and mapped the CpG island using the bioinformatic tool of CpG Plot. DNA was modified by the sodium bisulfite method, according to the EpiTect Bisulfite Kit (QIAGEN) instructions. Briefly, DNA was incubated at high concentrations of bisulfite salt at high temperature and low pH. A DNA-protecting buffer containing the pH indicator dye was used to confirm the correct pH for cytosine conversion.
The EpiTect Bisulfite procedure comprises a few simple steps: bisulfate-mediated conversion of unmethylated cytosine’s; binding of the converted single stranded DNA to the membrane of an EpiTect spin column; washing; desulfonation of membrane-bound DNA; washing of the membrane bound DNA to remove desulfonation agent; and elution of the pure, converted DNA from the spin column. Eluted DNA was dissolved in elution buffer (EB) and stored at -20ᵒC for future use.
D. Quantitative methylation-specific PCR (qMSP)
The methylation status of CpG sites within a CpG island was investigated using methylation-specific PCR (MSP). In this method, changes in DNA sequence after treatment with bisulfite in hypermethylated alleles are detected by converting methylated cytosine to uracil.
Subsequently PCR was done using primers specific to either methylated or unmethylated DNA. One microgram of DNA from each of 12 tissue sample and 12 blood samples of esophageal cancer were subjected to bisulfite treatment using the Epitect Bisulfite kit(Qiagen), following the manufacturer’s recommended procedures. RIZ1 promoter methylation was quantitatively assessed by qMSP assay. Real-time PCR reactions for RIZ1 gene was conducted with bisulfite modified DNA in 7900 HT (Applied Biosystems) instrument, using Power SYBR Green Supermix (Thermo Scientific) in a total reaction volume of 10 μl containing 1µg bisulfite treated DNA, using the primers as listed below. Beta-actin (ACTB) gene was regarded as an endogenous control where a region devoid of CpG islands was amplified. To ensure high reproducibility, RT-PCR reactions were repeated three times for each sample and negative DNA controls without template DNA were used in each RT-PCR test. The PCR program included initial denaturation at 95°C for 15 min followed by 40 cycles of 94°C for 15s and 55°C for 30s and 70°C for 34s.
The expression values of each gene were compared with the expression values of the reference gene to determine the DNA levels of the methylated promoter and the ratio obtained for easy tabulation was multiplied by 1000.
The data were analyzed using students’t-test (http:// www.graphpad.com/quickcalcs/) to compare the methylation status in tissue and blood samples. P values < 0.05 were considered to be statistically significant.
Results
A. Gene expression study
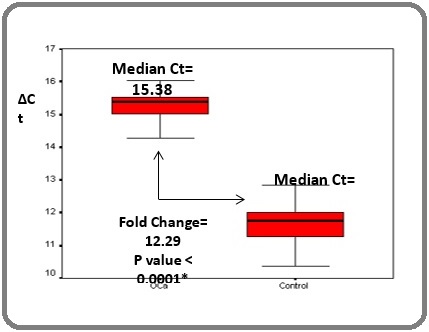
Quantitative gene expression of RIZ1 was studied in peripheral blood mononuclear cell (PBMC) fraction of esophageal cancer patients (n=15) and compared to that of healthy controls (n=12). A 12 fold down regulation (p<0.05) of RIZ1 in esophageal cancer (median delta Ct: 15.38) compared to the healthy controls (median delta Ct: 11.76) was observed (Figure 1).
Figure 1. Comparison of Relative Expression of RIZ1 Gene in the PBMC Fraction of Patients and Healthy Controls.
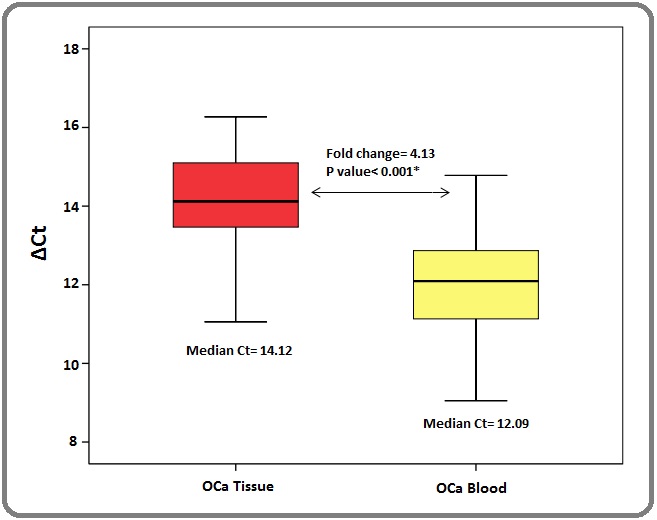
Similarly, comparative gene expression analysis for RIZ1 was performed between the tissue and blood level of EC patients (n=12). A 4.3 fold down regulation (p> 0.05) of RIZ1 expression in the tissue samples (median delta Ct : 14.12) of EC patients compared to their blood samples (median delta Ct : 12.09) was noted (Figure 2).
Figure 2. Comparison of Relative Expression of RIZ1 Gene between the Paired Samples.
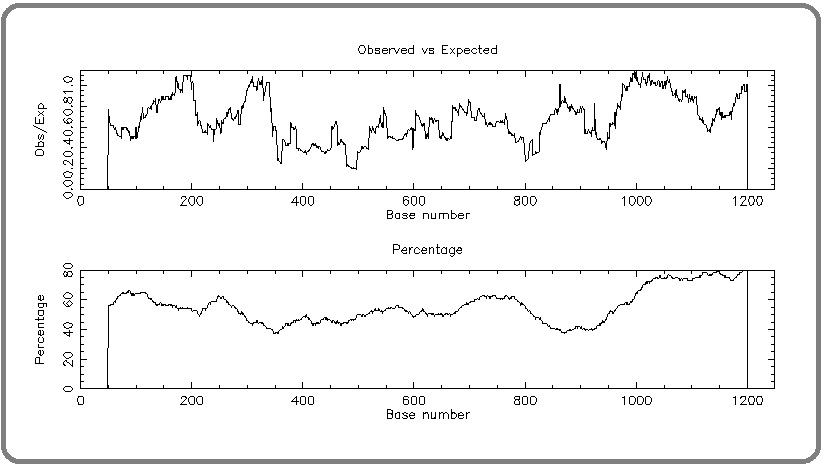
To address whether the deregulated expression of RIZ1 in a tissue-specific manner in cancer patients was attributed to any epigenetic factors, we moved on to study the promoter hyper-methylation status of RIZ1 gene at CpG dinucleotides. A map of the putative CpG islands in the promoter region of RIZ1 promoter was generated using CpG plot showing relatively higher density of CpG dinucleotides in the RIZ1 promoter (Figure 3).
Figure 3. A Map of the Putative CpG Islands in the Promoter Region of RIZ1 Promoter was Generated using CpG Plot Showing Relatively Higher Density of CpG Dinucleotides in the RIZ1 Promoter.
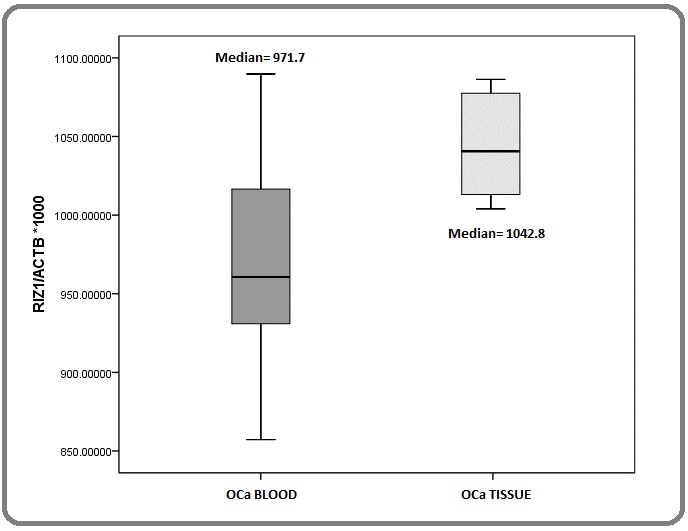
A quantitative estimation of methylation status of RIZ1 promoter was evaluated by quantitative methylation-specific PCR using bisulfite modified genomic DNA isolated from tissue and blood samples of EC patients.
The distribution of methylation quotients for blood and tissue samples did not differ significantly in EC patients rendering the role of promoter hypermethylation in altered gene expression of RIZ1 inconclusive in our patients (Figure 4).
Figure 4. Comparison of Methylation Quotients of the Specified Region of RIZ1 Promoter in the Genomic DNA Isolated from Eosophagal Tissues and Whole Blood.
Discussion
The present study a reduced RIZ-1 gene expression was documented in the patient’s as compared to the normal subjects (unpaired subjects) and the down regulation was 11.6 fold which was statistically significant). This indicates that in advanced or metastatic disease the reduction of RIZ1 expression could be one of the probable indicators of the metastatic state. When the RIZ-1 expression was compared in paired samples (the same person) A 4.28-fold downregulation of RIZ1 expression was detected.
Hypermethylation RIZ-1 promoter CpG island has been documented for a number of cancers. For example, RIZ1 hypermethylation has been reported in 42.6% of prostate cancer cases and was higher in patients with advanced stage of cancer. In thyroid carcinoma Lal et al (Lal G et al., 2006) reported that 100% cancerous cases were methylated, and methylation was significantly frequent compared with normal thyroid tissues (33%).
In gastric adenocarcinoma, Hypermethylation of this gene was reported in 69% of cancer tissues. However, in thyroid cancer, methylation of this gene was reported in 100% of cancer cases compared with 33% in normal thyroid tissue. The RIZ1 hypermethylation was identified in 44% (11/25) cases of breast cancer and in liver cancer samples it was 62%.
In modern day practice of clinical oncology, the focus is being gradually shifted from treating cancer patients with same histology and stage with emperic chemotherapy towards classifying the patient pool to different subsets based on molecular profile and treating each subset with a different and more tailored and customized chemotherapy based on their mode of action.
Hypermethylation causes gene silencing of tumor suppressor genes. This is reversible with drugs DNA Methyl tranferase inhibitors (DMTI) like 5 Azacytadine, Decitabine etc. These have not been used in EC so far. Our study aims to explore if differential methylation profile also if present in the EC patient pool.
In the present study, we conducted a quantitative methylation assay to compare the methylation status of RIZ-1 gene promoter in PBMC cells and cancer tissues to find out if the tissue specific downregulation of RIZ-1 gene is mediated through epigenetic factors- promoter CpG island hypermethylation. However, we could not detect any difference in the observed methylation quotients for RIZ-1 gene between PBMC and tissue samples. The probable reasons could be attributed to multiple factors. First, it can be said that in advanced EC, where the cancer is metastatic and the disease becomes systemic (stage IV is a systemic disease), status in methylation profile between different tissues gradually becomes uniform. Altered methylation may be prominent in early stages like in lung cancer. Secondly, the method applied here is dependent on the methylation status of the CpG dinucleotides that overlap with the primer sequences on the target template. The methylation density of the CpG islands internal to the primers was unaccounted here and this may vary among tissues. And finally, we have conducted this study with a limited number of patient samples, which might influence the observed inconclusiveness of the data.
However, our study unequivocally demonstrated a significant down-regulation of RIZ-1 gene in in the blood and tissue samples of EC patients from Indian origin, which has not been reported earlier.
In conclusion, our study demonstrated a significant down-regulation of RIZ-1 gene in the blood and tissue samples of EC patients from Indian origin, which has not been reported earlier. We also observed that the altered promoter methylation was not related the observed down-regulation among patient’s higher stage.
References
- Overexpression of cyclin B1 in human esophageal squamous cell carcinoma cells induces tumor cell invasive growth and metastasis Song Yongmei, Zhao Chunling, Dong Lijia, Fu Ming, Xue Liyan, Huang Zhen, Tong Tong, Zhou Zhuan, Chen Amei, Yang Zhihua, Lu Ning, Zhan Qimin. Carcinogenesis.2007;29(2). CrossRef
- Prognostic significance of PTEN expression in esophageal squamous cell carcinoma from Linzhou City, a high incidence area of northern China Chang D., Wang T.-Y., Li H.-C., Wei J.-C., Song J.-X.. Diseases of the Esophagus.2007;20(6). CrossRef
- Etiology and chemoprevention of esophageal squamous cell carcinoma Stoner Gary D., Gupta Ashok. Carcinogenesis.2001;22(11). CrossRef
- EMP3 as a tumor suppressor gene for esophageal squamous cell carcinoma Noguchi Tsuyoshi, Nishiyama Masahiko. Cancer Letters.2009;274(1). CrossRef
- Decreased expression of RUNX3 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma Sugiura H, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, et al . Oncol Rep.2008;19(3):713-719.
- The Clinical Significance of Aurora-A/STK15/BTAK Expression in Human Esophageal Squamous Cell Carcinoma Tanaka Eiji, Hashimoto Yosuke, Ito Tetsuo, Okumura Tomoyuki, Kan Takatsugu, Watanabe Go, Imamura Masayuki, Inazawa Johji, Shimada Yutaka. Clinical Cancer Research.2005;11(5). CrossRef
- EMP3 as a tumor suppressor gene for esophageal squamous cell carcinoma Fumoto Shoichi, Hiyama Keiko, Tanimoto Keiji, Noguchi Takuya, Hihara Jun, Hiyama Eiso, Noguchi Tsuyoshi, Nishiyama Masahiko. Cancer Letters.2009;274(1). CrossRef
- Inverse expression of dihydrodiol dehydrogenase and glutathione-S-transferase in patients with esophageal squamous cell carcinoma Wang Liang-Shun, Chow Kuan-Chih, Wu Yu-Chung, Lin Tze-Yi, Li Wing-Yin. International Journal of Cancer.2004;111(2). CrossRef
- survivin messenger RNA expression is a good prognostic biomarker for oesophageal carcinoma Ikeguchi M, Kaibara N. British Journal of Cancer.2002;87(8). CrossRef
- Osteopontin Expression in Squamous Cell Cancer of the Esophagus Wu I-Chen, Wu Ming-Tsang, Chou Shah-Hwa, Yang Sheau-Fang, Goan Yih-Gang, Lee Jang-Ming, Chou Yi-Pin, Bair Ming-Jong, Wang Tsang-En, Chen Angela, Chang Wen-Hsiung, Kuo Fu-Chen, Wu Deng-Chyang. World Journal of Surgery.2008;32(9). CrossRef
- Discovery of deregulation of zinc homeostasis and its associated genes in esophageal squamous cell carcinoma using cDNA microarray Kumar Anupam, Chatopadhyay Tusharkant, Raziuddin Mohamad, Ralhan Ranju. International Journal of Cancer.2006;120(2). CrossRef
- Gene expression profile of esophageal cancer in North East India by cDNA microarray analysis Chattopadhyay Indranil. World Journal of Gastroenterology.2007;13(9). CrossRef
- Genome-wide analysis of chromosomal alterations in patients with esophageal squamous cell carcinoma exposed to tobacco and betel quid from high-risk area in India Chattopadhyay Indranil, Singh Avninder, Phukan Rupkumar, Purkayastha Joydeep, Kataki Amal, Mahanta Jagadish, Saxena Sunita, Kapur Sujala. Mutation Research/Genetic Toxicology and Environmental Mutagenesis.2010;696(2). CrossRef
- Contribution of germ lineBRCA2sequence alterations to risk of familial esophageal cancer in a high-risk area of India Kaushal M., Chattopadhyay I., Phukan R., Purkayastha J., Mahanta J., Kapur S., Saxena S.. Diseases of the Esophagus.2010;23(1). CrossRef
- Role of epoxide hydrolase 1 gene polymorphisms in esophageal cancer in a high-risk area in India Ihsan Rakhshan, Chattopadhyay Indranil, Phukan Rupkumar, Mishra Ashwani K, Purkayastha Joydeep, Sharma Jagannath, Zomawia Eric, Verma Yogesh, Mahanta Jagadish, Saxena Sunita, Kapur Sujala. Journal of Gastroenterology and Hepatology.2010;25(8). CrossRef
- CASP8 −652 6N del and CASP8 IVS12-19G>A gene polymorphisms and susceptibility/prognosis of ESCC: A case control study in northern Indian population Umar Meenakshi, Upadhyay Rohit, Kumar Shaleen, Ghoshal Uday Chand, Mittal Balraj. Journal of Surgical Oncology.2011;103(7). CrossRef
- Association of Xenobiotic Metabolizing Enzymes Genetic Polymorphisms With Esophageal Cancer in Kashmir Valley and Influence of Environmental Factors Malik Manzoor Ahmad, Upadhyay Rohit, Mittal Rama Devi, Zargar Showket Ali, Mittal Balraj. Nutrition and Cancer.2010;62(6). CrossRef
- Functional polymorphisms of cyclooxygenase-2 (COX-2) gene and risk for esophageal squmaous cell carcinoma Upadhyay Rohit, Jain Meenu, Kumar Shaleen, Ghoshal Uday Chand, Mittal Balraj. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis.2009;663(1-2). CrossRef
- Esophageal Cancer Enzinger Peter C., Mayer Robert J.. New England Journal of Medicine.2003;349(23). CrossRef
- Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, et al . Cancer: Interdisciplinary International Journal of the American Cancer Society.2001;92(11):2769-2777. CrossRef
- Genetic analysis of candidate genes modifying the age-at-onset in Huntington’s disease Metzger Silke, Bauer Peter, Tomiuk Jürgen, Laccone Franco, DiDonato Stefano, Gellera Cinzia, Mariotti Caterina, Lange Herwig W., Weirich-Schwaiger Helga, Wenning Gregor K., Seppi Klaus, Melegh Bela, Havasi Viktoria, Balikó Laszlo, Wieczorek Stefan, Zaremba Jacek, Hoffman-Zacharska Dorota, Sulek Anna, Basak A. Nazli, Soydan Esra, Zidovska Jana, Kebrdlova Vera, Pandolfo Massimo, Ribaï Pascale, Kadasi Ludovit, Kvasnicova Marta, Weber Bernhard H. F., Kreuz Friedmar, Dose Matthias, Stuhrmann Manfred, Riess Olaf. Human Genetics.2006;120(2). CrossRef
- The Retinoblastoma Interacting Zinc Finger Gene RIZ Produces a PR Domain-lacking Product through an Internal Promoter Liu Limin, Shao Gang, Steele-Perkins George, Huang Shi. Journal of Biological Chemistry.1997;272(5). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details