Profile of PD-L1 mRNA Expression in Childhood Acute Leukemia
Download
Abstract
Background: Programmed Death Ligand 1 (PD-L1) expression was used to determine type of patients who respond to immunotherapy using immune checkpoint inhibitor in several solid malignancies. However, the role of PD-L1 in hematological malignancies is less explored. Therefore, we aim to investigate PD-L1 mRNA expression in childhood leukemia patients. Association between PD-L1 expression and clinicopathological features was also analyzed.
Method: Blood samples of 17 childhood leukemia patients and 12 healthy individuals as control were used in this study. The samples were collected in Dharmais Cancer Hospital. Real time PCR was used to analyze PD-L1 expression with GAPDH as internal control.
Result: PD-L1 mRNA expression is significantly lower in childhood leukemia patients (medians: 0.0012) compared to healthy individuals (medians: 0.0041) (p=0.017). PD-L1 mRNA expressions were not correlated with age (p=1.000), sex (p=1.000), outcome (p=1.000), type of leukemia (p=1.000), hyperleukocytosis (p=1.000), and syndrome down (p=0.426).
Conclusion: PD-L1 expression is lower in leukemia patients compared to healthy controls. PD-L1 expressions were not correlated with all clinicopathological features. Our result provides preliminary data of PD-L1 expression in Indonesian childhood leukemia patients.
Introduction
Leukemia is most common cancer in children with high mortality rate. Acute Lymphocytic Leukemia (ALL) and Acute Myelogenous Leukemia (AML) are 2 major types in pediatric leukemia [1]. Survival rates have improved in children with leukemia due to advances in treatment and supportive care [2]. However, rate of survival in childhood leukemia is still low in developing countries including in Indonesia [3]. Standard therapy for children with leukemia such as radiation and chemotherapy is not effective enough especially for those who faced resistant or recurrent cancers [4-5]. Patients with refractory, high-grade, recurrent or metastatic disease are still undergone extremely poor outcomes [2]. Therefore, novel therapy to treat childhood cancer is needed to improve patient’s outcome.
Immunotherapy has emerged as an effective cancer treatment. CAR T cell therapy is one type of immunotherapy that has shown promising result in treating childhood leukemia [4]. In solid tumor, immune checkpoint inhibitors such as anti-PD-1 and anti-PD-L1 have been approved by Food and Drug Administration (FDA) to treat several cancers such as melanoma and non-small cell lung cancer (NSCLC) [6]. Anti-PD-1/ PD-L1treatment is type of immunotherapy that use monoclonal antibody (mAb) to inhibit the binding between programmed cell death-1 (PD-1) and programmed cell death ligand-1 (PD-L1) molecules in cancer patients [7-8]. Binding interaction between PD-1 and PD-L1 significantly down regulates T cell, thus causes tumor growth, promotes metastasis, and mediates immune suppression [8-10]. Therefore, blocking the engagement between these 2 molecules could enhance T cell responses and increase anti-tumor immunity [8-11]. In childhood cancer, pembrolizumab, mAb for PD-1 has been approved by FDA for the treatment of children with refractory classic Hodgkin Lymphoma (HL) [4]. In childhood leukemia, clinical trials using immune checkpoint inhibitors is currently on going [2]. A case report suggested that pediatric patient with multiple relapsed and refractory acute leukemia treated with combination of 5-azacytidine with immune checkpoint inhibitors (nivolumab and ipilimumab) is tolerated the therapy and showed improvement of symptoms [12]. It has been suggested that combinations of multiple immunotherapeutic and conventional therapy such as chemotherapy might improve pediatric cancer outcome [2-4]. Currently, there is an on-going clinical trial (NCT02813135) for combination of cyclophosphamide and nivolumab for the treatment of recurrent pediatric cancers [4].
PD-1 is an inhibitory receptor expressed on activated T cell. PD-L1 is a PD-1’s ligand and commonly expressed on cancer cells and immune cells like T cell and B cell [8]. PD-L1 has 40-kDa size and categorized as type 1 trans-membrane cell surface glycoprotein and member of the B7 family of co-stimulatory molecules [2-9-13]. Studies have shown that expression of PD-L1 is associated with poor prognosis in several cancers such as melanoma and non-small cell lung cancer (NSCLC) including in pediatric cancer [2-4-14]. PD-L1 expression also has been correlated with response to anti PD-1/ PD-L1 therapy [14]. In fact, PD-L1 expression has been used to determine type of patients who respond to immune checkpoint inhibitors in several cancers [6]. FDA has approved the measurement of PD-L1 expression using immunohistochemistry as companion diagnostic for the treatment of NSCLC and melanoma using anti PD-1 [6-8].
However, in hematological malignancies especially in children, the role of PD-L1 is less explored. There are only a very few studies investigating PD-L1 expression in pediatric malignancies [15]. Besides, to the best of our knowledge, there is no study analyzing PD-L1 in childhood leukemia that has been done in Indonesia. Therefore, we aim to analyze PD-L1 mRNA expression in blood samples of childhood leukemia patients in Dharmais Cancer Hospital (National Cancer Center of Indonesia) and investigate the relationship between PD-L1 expression and clinicopathological features.
Materials and Methods
Samples Collection
We examined 17 blood samples of childhood leukemia patients which consist of 11 samples of ALL patients and 6 samples of AML patients. All cases have been confirmed in the Clinical Pathology Department, Dharmais Cancer Hospital. The 17 patients included 9 males and 8 females with an age ranging from 2 to 15 years (mean ages: 6.1±4.2). As many as 12 healthy people were used as controls that were consisted of 6 males and 6 females. Blood samples were collected in ethylenediaminetetraacetic acid (EDTA) tubes then directly processed for RNA isolation. This study has been approved by Ethical Committee of Dharmais Cancer Hospital (code: 065/KEPK/VI/2020).
Real time PCR analysis
RNA molecule was extracted using Total RNA Blood/ Culture (Gene Aid). A maximum of 2000 ng of RNA was then reverse transcribed to cDNA using High Capacity cDNA Sythesis Kit (Applied Biosystem). Dilution to 100 ng was done to all cDNA samples. Diluted cDNA samples were then used in real time PCR analysis.
To avoid genomic DNA amplification, primers used in real time PCR were designed to span 2 different exons. For PD-L1, we used forward primer 5’-tgtgaaagtcaatgccccat-3’, reverse primer 5’-tgtcagttcatgttcagaggt-3’, and probe 5’-attttggttgtggatccagtc-3’. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. For GAPDH, the forward primer, reverse primer, and probe were 5’-agcctcaagatcatcagcaa-3’, 5’-actgtggtcatgagtccttc-3’, and 5’-ctgcaccaccaactgcttag-3’, respectively. All primers and probes were formulated and mixed by Applied Biosystem into Custom TaqMan Gene Expression Assay.
Single real time PCR reaction consisted of 10 µL of TaqMan Gene Expression Master Mix (Applied Biosystem), 1 µL of Custom TaqMan Gene Expression Assay (Applied Biosystem), 5 µL of Nuclease Free Water, and 4 µL of cDNA. We doubled the reaction for each sample. The thermal cycling condition for qPCR were: 2 minutes (50oC) and 10 minutes (95oC) hold stage, followed by 40 cycles of denaturation step at 95oC for 15 seconds, and annealing and extension step at 62oC for 1 minute. The reaction was performed in 7500 Fast Real-Time PCR System (Applied Biosystem).
Data was analyzed using 2-∆∆CT (∆∆CT = ∆CT of each samples – average ∆CT of healthy controls) to get the value of gene expression. We classified the level of gene expression as low or high expression by using average ΔCT value of 12 healthy people as cut-off value. ∆CT values of samples below average ∆CT value of healthy controls were determined as high expression. Meanwhile, samples with ∆CT values above average ∆CT value of healthy controls were determined as low expression.
Statistical analysis
Statistical analysis was done using SPSS Statistics 22 statistical software (IBM Corporation). Fisher exact test was used to analyze the relationship between PD-L1 expression and clinicopathological characteristics. To analyze the comparison of PD-L1 expression in cancer and healthy control group, Man-Whitney test was performed. A value of P<0.05 is considered statistically significant.
Results
The expression of PD-L1 in childhood leukemia patients
PD-L1 expression in childhood leukemia patients is significantly lower compared to healthy controls. Median expression of PD-L1 in childhood leukemia is 0.0012 (ranges from 0.00005 to 0.077), while median expression of PD-L1 in healthy control group is 0.0041 (ranges from 0.0016 to 0.013) (p=0.017) (Figure 1).
Figure 1: PD-L1 Gene Expression in Childhood Leukemia (Leukemia) and Healthy Controls (HC).
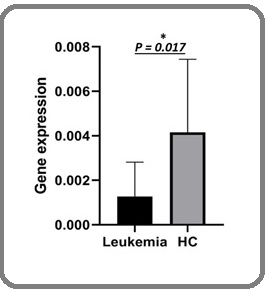
Gene expression in figure 1 is the result of 2-∆∆CT calculation method with median and error bar (95% confidence interval).
The association of PD-L 1 expression and clinicopathological characteristic of childhood leukemia patients
From 17 samples analyzed, as many as 4 patients were categorized as high expression, while 13 patients were categorized as low expression. The expression of PD-L1 in childhood leukemia patients was not associated with all clinicopathological characteristics examined. The result showed that there was no significant relationship between PD-L1 mRNA expressions with age (p=1.000), sex (p=1.000), outcome (p=1.000), type of leukemia (p=1.000), hyperleukocytosis (p=1.000), and syndrome down (p=0.426) (Table 1).
| Parameter | Low expression | High expression | p-value | |
| n (%) | n (%) | |||
| Age | ≤10 | 11 (78.6) | 3 (21.4) | 1 |
| >10 | 2 (66.7) | 1 (33.3) | ||
| Sex | Male | 7 (77.8) | 2 (22.2) | 1 |
| Female | 6 (75) | 2 (25) | ||
| Type of Leukemia | ALL | 8 (72.8) | 3 (27.2) | 1 |
| AML | 5 (83.3) | 1 (16.7) | ||
| Syndrome Down | Yes | 1 (50) | 1 (50) | 0.426 |
| No | 12 (80) | 3 (20) | ||
| Hyperleukocytosis | Yes | 2 (100) | 0 (0) | 1 |
| No | 11 (73.3) | 4 (26.7) | ||
| Outcome | Life | 11(78.6) | 3 (21.4) | 1 |
| Dead | 2 (66.7) | 1 (33.3) |
AML, Acute Myelogenous Leukemia; ALL, Acute Lymphocytic Leukemia
Discussion
This study revealed that PD-L1 expression is significantly lower in childhood leukemia patients compared to healthy controls. We found a low frequency of PD-L1 high expression in pediatric leukemia. Only 23.5% (4/17) patients of childhood leukemia showed high PD-L1 expression. The same finding also found in previous reports in pediatric solid tumors. Positive PD-L1 expression was only found in 11 of 81 (14%) of Wilms tumors [16] and only 13% of total 500 cases of pediatric tumors (including neuroblastoma, brain cancers, and sarcomas) [17]. Another study by Aoki et al. [18] showed positive PD-L1 expression only found in 1 sample of rhabdomyosarcoma out of 53 samples of pediatric solid tumors (8 extracranial malignant germ cell tumors, 18 neuroblastomas, 7 hepatoblastomas, 7 germinomas, 4 medulloblastomas, 4 renal tumors, 2 atypical teratoid/ rhabdoid tumor (AT/RT), and 3 rhabdomyosarcomas).
In human adults with leukemia, Chen et al. [19] described positive PD-L1 expressions were higher in monocyte leukemia (M5) patients than other type of leukemia (24%), while Yang et al. [20] showed that PD-L1 mRNA expression were significantly higher in T-Acute lymphoblastic leukemia (T-ALL) compared to reactive hyperplasia. Meanwhile, in childhood leukemia, study by Feucht et al. [21] revealed inter-individual differences in PD-L1 expressions of childhood ALL. Variable PD-L1 expression patterns were found on patients’ bone marrow blasts [21], suggested that PD-L1 expressions in childhood leukemia could be highly vary among different individuals. Moreover, Feucht et al. [21] revealed that high PD-L1 expression was mostly found in pediatric ALL patients who relapsed and not respond to Blinatumomab treatment.
It has been explained that PD-L1 gene abnormality such as change in PD-L1 gene copy number status might contribute to the increase of PD-L1 expression in cancer. Study by Inoue et al. [22] showed that tumor with PD-L1 genomic gains exhibit significantly higher PD-L1 expression compared to those without. Moreover, it has been shown that pediatric cancer seems to have lower gene mutation burden compared to cancer in adults [2-4]. In general, childhood cancers do not have high mutations rate [23]. Because of the low level of gene mutation burden in pediatric cancer, PD-L1 gene abnormality might not present in childhood leukemia, thus diminish one of the factors that is responsible for enhancing PD-L1 expression. This could explain why PD-L1 expression is lower in childhood leukemia. However, it should be confirmed with further study to analyze the correlation between PD-L1 gene abnormalities with PD-L1 expression in childhood leukemia.
This study also found that PD-L1 expression is not associated with all the clinicopathological parameters such as age, sex, outcomes, type of leukemia, hyperleukocytosis, and syndrome down. This might be due to the small sample size in our study. To find significant correlation, number of samples need to be added.
Pediatric cancer patients tend to exhibit poor response toward single immune checkpoint inhibitor therapy (anti-PD-1/ anti PD-L1) [4]. Low tumor mutation load might be one of the factors that cause poor response rate of anti-PD-1/ anti PD-L1 therapy in pediatric cancer [4]. In this study, we found most childhood leukemia patients have low PD-L1 expression. Since low PD-L1 expression is mostly related with poor clinical outcomes to anti-PD-1-directed therapy [24], this result might additionally explain why pediatric cancer patients seem to not give good response to single immune checkpoint inhibitor. It has been suggested that combination of therapy such as immune checkpoint inhibitor and chemotherapy might enhance response rate of pediatric cancer and improve survival [2-4]. However, the limitation of this study is very small sample size. To obtain more accurate data, more samples need to be added and comparison with immunohistochemistry of PD-L1 as gold standard to measure PD-L1 expression need to be done in the subsequent experiment.
In conclusion, PD-L1 expression seems to have low expression level in childhood leukemia. This might explain why single immune checkpoint inhibitor therapy did not show very good result in pediatric cancer patients. Besides, we also found, the expressions of PD-L1 were not correlated with all clinicopathological features. This result provides preliminary data of PD-L1 expression in Indonesian pediatric leukemia patients.
Acknowledgements
The authors wish to thank to Department of Research and Development, Dharmais Hospital-National Cancer Center for providing the funding and instrument used in the study.
Competing of Interest
The authors declare no conflict of interest.
References
- Pediatric leukemia : Diagnosis to treatment - A review Bernard SC, Abdelsamad EH, Johnson PA, Chapman DL, Parvathaneni M. Journal of Cancer Clinical trials.2017;2(2):2-4.
- Pediatric Malignancies [Internet]. Immune Checkpoint Inhibitors in Cancer Ring EK, Gillespie GY, Friedman GK. Elsevier Inc.;:193-204.
- The Challenge of Childhood Cancer in Developing Countries Haileamlak A. Ethiopian journal of health sciences.2016;26(3):199-200.
- Pediatric Cancer Immunotherapy: Opportunities and Challenges Wedekind Mary Frances, Denton Nicholas L., Chen Chun-Yu, Cripe Timothy P.. Pediatric Drugs.2018;20(5). CrossRef
- Childhood and adolescent cancer statistics, 2014 Ward Elizabeth, DeSantis Carol, Robbins Anthony, Kohler Betsy, Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2014;64(2). CrossRef
- Optimization and validation of PD-L1 immunohistochemistry staining protocols using the antibody clone 28-8 on different staining platforms Koppel Christina, Schwellenbach Helena, Zielinski Dirk, Eckstein Sina, Martin-Ortega Mercedes, D’Arrigo Corrado, Schildhaus Hans-Ulrich, Rüschoff Josef, Jasani Bharat. Modern Pathology.2018;31(11). CrossRef
- The blockade of immune checkpoints in cancer immunotherapy Pardoll Drew M.. Nature Reviews Cancer.2012;12(4). CrossRef
- Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy Topalian Suzanne L., Taube Janis M., Anders Robert A., Pardoll Drew M.. Nature Reviews Cancer.2016;16(5). CrossRef
- Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion Dong Haidong, Strome Scott E., Salomao Diva R., Tamura Hideto, Hirano Fumiya, Flies Dallas B., Roche Patrick C., Lu Jun, Zhu Gefeng, Tamada Koji, Lennon Vanda A., Celis Esteban, Chen Lieping. Nature Medicine.2002;8(8). CrossRef
- Anti–PD-1/PD-L1 therapy of human cancer: past, present, and future Chen Lieping, Han Xue. Journal of Clinical Investigation.2015;125(9). CrossRef
- Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., Minato N.. Proceedings of the National Academy of Sciences.2002;99(19). CrossRef
- Checkpoint inhibition of PD-L1 and CTLA-4 in a child with refractory acute leukemia Broglie Larisa, Gershan Jill, Burke Michael J. International Journal of Hematologic Oncology.2019;8(1). CrossRef
- Tissue expression of PD-L1 mediates peripheral T cell tolerance Keir Mary E., Liang Spencer C., Guleria Indira, Latchman Yvette E., Qipo Andi, Albacker Lee A., Koulmanda Maria, Freeman Gordon J., Sayegh Mohamed H., Sharpe Arlene H.. Journal of Experimental Medicine.2006;203(4). CrossRef
- Predictive biomarkers for checkpoint inhibitor-based immunotherapy Gibney Geoffrey T, Weiner Louis M, Atkins Michael B. The Lancet Oncology.2016;17(12). CrossRef
- The PD-L1/PD-1 axis expression on tumor-infiltrating immune cells and tumor cells in pediatric rhabdomyosarcoma Gabrych Anna, Pęksa Rafał, Kunc Michał, Krawczyk Małgorzata, Izycka-Swieszewska Ewa, Biernat Wojciech, Bień Ewa. Pathology - Research and Practice.2019;215(12). CrossRef
- B7-H1 Expression in Wilms Tumor: Correlation With Tumor Biology and Disease Recurrence Routh Jonathan C., Ashley Richard A., Sebo Thomas J., Lohse Christine M., Husmann Douglas A., Kramer Stephen A., Kwon Eugene D.. Journal of Urology.2008;179(5). CrossRef
- Programmed Death-Ligand 1 Expression in a Large Cohort of Pediatric Patients With Solid Tumor and Association With Clinicopathologic Features in Neuroblastoma Saletta Federica, Vilain Ricardo E., Gupta Aditya Kumar, Nagabushan Sumanth, Yuksel Aysen, Catchpoole Daniel, Scolyer Richard A., Byrne Jennifer A., McCowage Geoffrey. JCO Precision Oncology.2017;(1). CrossRef
- Low Frequency of Programmed Death Ligand 1 Expression in Pediatric Cancers Aoki Takahiro, Hino Moeko, Koh Katsuyoshi, Kyushiki Masashi, Kishimoto Hiroshi, Arakawa Yuki, Hanada Ryoji, Kawashima Hiroshi, Kurihara Jun, Shimojo Naoki, Motohashi Shinichiro. Pediatric Blood & Cancer.2016;63(8). CrossRef
- Clinical significance of B7-H1(PD-L1)expression in human acute leukemia Chen Xiangli, Liu Shuhu, Wang Liancai, Zhang Wang-Gang, Ji Yuqiang, Ma Xiaorong. Cancer Biology & Therapy.2008;7(5). CrossRef
- Expression and significance of CD47, PD1 and PDL1 in T-cell acute lymphoblastic lymphoma/leukemia Yang Kun, Xu Jing, Liu Qinghang, Li Jing, Xi Yanfeng. Pathology - Research and Practice.2019;215(2). CrossRef
- T-cell responses against CD19+ pediatric acute lymphoblastic leukemia mediated by bispecific T-cell engager (BiTE) are regulated contrarily by PD-L1 and CD80/CD86 on leukemic blasts Feucht Judith, Kayser Simone, Gorodezki David, Hamieh Mohamad, Döring Michaela, Blaeschke Franziska, Schlegel Patrick, Bösmüller Hans, Quintanilla-Fend Leticia, Ebinger Martin, Lang Peter, Handgretinger Rupert, Feuchtinger Tobias. Oncotarget.2016;7(47). CrossRef
- Clinical significance of PD-L1 and PD-L2 copy number gains in non-small-cell lung cancer Inoue Yusuke, Yoshimura Katsuhiro, Mori Kazutaka, Kurabe Nobuya, Kahyo Tomoaki, Mori Hiroki, Kawase Akikazu, Tanahashi Masayuki, Ogawa Hiroshi, Inui Naoki, Funai Kazuhito, Shinmura Kazuya, Niwa Hiroshi, Suda Takafumi, Sugimura Haruhiko. Oncotarget.2016;7(22). CrossRef
- Mutational heterogeneity in cancer and the search for new cancer-associated genes Lawrence Michael S., Stojanov Petar, Polak Paz, Kryukov Gregory V., Cibulskis Kristian, Sivachenko Andrey, Carter Scott L., Stewart Chip, Mermel Craig H., Roberts Steven A., Kiezun Adam, Hammerman Peter S., McKenna Aaron, Drier Yotam, Zou Lihua, Ramos Alex H., Pugh Trevor J., Stransky Nicolas, Helman Elena, Kim Jaegil, Sougnez Carrie, Ambrogio Lauren, Nickerson Elizabeth, Shefler Erica, Cortés Maria L., Auclair Daniel, Saksena Gordon, Voet Douglas, Noble Michael, DiCara Daniel, Lin Pei, Lichtenstein Lee, Heiman David I., Fennell Timothy, Imielinski Marcin, Hernandez Bryan, Hodis Eran, Baca Sylvan, Dulak Austin M., Lohr Jens, Landau Dan-Avi, Wu Catherine J., Melendez-Zajgla Jorge, Hidalgo-Miranda Alfredo, Koren Amnon, McCarroll Steven A., Mora Jaume, Lee Ryan S., Crompton Brian, Onofrio Robert, Parkin Melissa, Winckler Wendy, Ardlie Kristin, Gabriel Stacey B., Roberts Charles W. M., Biegel Jaclyn A., Stegmaier Kimberly, Bass Adam J., Garraway Levi A., Meyerson Matthew, Golub Todd R., Gordenin Dmitry A., Sunyaev Shamil, Lander Eric S., Getz Gad. Nature.2013;499(7457). CrossRef
- PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy Patel Sandip Pravin, Kurzrock Razelle. Molecular Cancer Therapeutics.2015;14(4). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details