Prevalence of Human Cytomegalovirus in Breast Cancer: A Systematic Review
Download
Abstract
Background: Breast cancer is the most common malignancy among women around the world, with several risk factors contributing to its progression. The role of infectious agents such as viruses in the progression of this cancer is relatively well-known, as a result of various studies in different geographical regions that reported the prevalence of cytomegalovirus in breast cancer.
Objective: In this systematic review, we tried to examine the prevalence of cytomegalovirus in breast cancer.
Materials and Methods: We conducted a comprehensive search of three databases: PubMed, Scopus, and Google Scolar up to January 21, 2021. Results: Out of 584 articles, 29 articles including 3158 samples were finally reviewed, of which 16 articles used PCR methods, 8 articles used serology method and 13 articles used other methods such as immunohistochemistry and in situ hybridization. The prevalence of the virus was higher in Asian and African countries such as Iraq and Egypt than other European and South American countries. Among these, the highest prevalence of the virus in different samples of patients with breast cancer was related to Iraq.
Conclusion: The results of our study showed the higher prevalence of the virus in patients with breast cancer rather than normal subjects and reveal the role of the virus in the development of breast cancer.
Introduction
Malignancies are one of the leading causes of death in this century. According to evidence, 9.6 million people died of cancer only in 2018 worldwide that among them breast cancer with 626679 cases of death has been introduced as the most common cancer among women around the world [1]. However, this type of malignancy has a worldwide prevalence, its incidence, mortality, and survival rate vary significantly in different parts of the world [2]. An important point in increasing the incidence of breast cancer in communities may originate from risk factors variations [3]. Breast cancer can be associated with various factors such as demographic structures, lifestyle, genetic factors, environment and etc [2]. The incidence of breast cancer can be strongly related to race and ethnicity [4]. The incidence of this malignancy in different parts of the world varies from 27 per 100,000 people (Central Africa and East Asia) to 92 per 100,000 people (North America) [5]. Extensive researches from around the world have clarified that there is an almost 4-fold difference in the incidence of breast cancer between developed and undeveloped countries, which reveals the profound impact of environmental factors on the process of disease development [1]. As mentioned, there are several factors involved in the development of breast cancer malignancy, which according to the International Agency for Research on Cancer the biological carcinogens are one of the influential factors in this process [6]. Biological carcinogens are one of the most important factors involved in breast cancer promotion. Over the past decades, the hypothesis of the role of viruses in breast carcinogenesis has attracted extensive attention. The viruses including human papilloma viruses (HPVs), Epstein–Barr virus (EBV-also known as human herpes virus type 4), mouse mammary tumor virus (MMTV), bovine leukemia virus (BLV), and human cytomegalovirus (HCMV) have been mentioned to be related to breast cancer [7]. Although the exact role of these viruses in the breast cancer remains controversial, large body of studies reported an association between HCMV and breast cancer [8].
The HCMV is a member of the herpesviridae family that has the ability to cause latent infections among individuals [9]. This virus, like most members of the herpesviridae family, is highly prevalent in different communities, with prevalence rate of 50 to 100% around the world [9]. The features including targeting cancer-related factors Rb and p53, the ability of cellular transformation “in vitro”, encoding proteins with oncogenic functions such as IE and pp65, employing mechanisms of oncomodulation and evading the immune system responses, have turned HCMV into an influential factor involved in pathogenesis of human malignancies, such as glioma, neuroblastoma, and breast cancer [10-11]. Existing data has shown the presence of CMV in different samples of patients with breast cancer [8]. In this study, we tried to assess the prevalence of CMV in patients with breast cancer. We also aimed to highlight the prevalence rate of CMV in tissue and also its serum of patients’ prevalence in different regions of the world by comparing different methods.
Materials and Methods
Literature search strategy
This systematic review investigated the techniques including conventional Polymerase chain reaction (PCR), Nested-PCR, Real-time PCR, serological, immunohistochemistry, and hybridization for determining the prevalence of HCMV in patients with breast cancer. The PubMed, Scopus and Google Scholar databases were searched up to January 21, 2021 using combinations of the following key words: (Cytomegalovirus OR Cytomegalovirus infection OR CMV OR human herpesvirus-5 OR HCMV) and (breast cancer OR breast neoplasm OR breast tumor OR cancer of breast OR human mammary neoplasm OR human mammary carcinoma OR breast carcinoma).
Study selection
At first, the selected articles were entered into EndNote software version X8 from three databases and the duplicate items were removed, then by screening the titles and abstracts, a number of articles were also deleted. The remaining studies were reviewed by two reviewers based on established criteria.
Inclusion and exclusion criteria
Studies with pre-determined criteria were selected for the current study which include: a) All case-control and cross-sectional studies conducted based on the conventional PCR, Nested-PCR, Real-time PCR, serological, immunohistochemistry, and hybridization methods for determining the prevalence of HCMV in patients with different types of breast cancer, b) Studies examining fresh tissue or formalin-fixed paraffin embedded (FFPE) tissues, blood, plasma, serum c) Utilization of samples of adjacent normal tissue or healthy individuals without breast cancer, fibroadenoma and benign as a control group, d) Full text studies in English.
Exclusion criteria also include: a) Review studies, case reports, abstract, master’s thesis or doctoral thesis and letter to editor, b) Studies on cell culture and animal, c) Studies that have used benign tissues such as fibroadenoma, fibrocystic, mastitis as a case sample, d) Studies that have considered non-inflammatory breast cancer samples as controls, e) Samples of cancer or normal are related to men, f) Studies that have used diagnostic methods conventional PCR, Nested-PCR, Real-time PCR, serological, immunohistochemistry, and hybridization, g) Studies that have examined the effect of the drug on HCMV-related breast cancer, h) Studies published in languages other than English.
Data Extraction
For each included study, two authors reviewed and extracted following data: author name, year of study, country of study, type of study, sample size, type of cancer, type of specimen, type of method used and results of the study are reported numerically and as a percentage.
Ethical Review and Reporting
Given that this systematic review study used the results of others studies and did not use directly human samples or animal, therefore was not submitted for any ethical approval. This study is reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Results
Literature Search
The article selection flowchart is shown in Figure 1.
Figure 1. Flowchart of Systematic Literature Search and Article Selection.
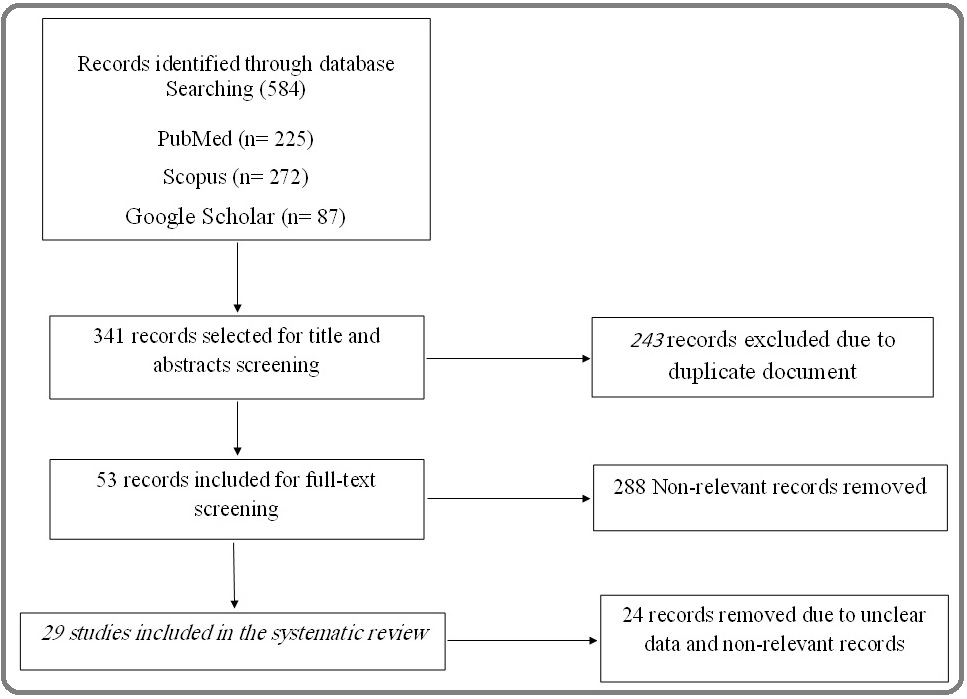
Initially, 584 articles were selected from three databases by searching for the relevant keywords, and after removing the duplicate items, 341 articles remained. 288 articles were excluded due to irrelevance to the subject by reviewing the titles and abstracts of the articles by three reviewers. 24 articles removed due to unclear data and non-relevant records. The remaining 29 articles were reviewed in full text, of which 16 articles used PCR methods, 8 articles used serological method, 9 articles used immunohistochemistry (IHC) method, 4 articles used in situ hybridization (ISH) techniques and 7 articles employed the mentioned techniques in combination. In addition, Tables 1 to 3 provides information such as techniques used, type of tissue studied, target gene studied, reporting countries, number of breast cancer cases, and cases of breast cancer and normal samples that were positive for the CMV.
| Gene | Type of Tissue | Test | Studies | Cases | Controls | Significant Correlation | ||||
| No. | No. | Positivity | No. | No. | Positivity | |||||
| Positive | Tested | Positive | Tested | |||||||
| IE | FFPE | PCR | Aidrous et al, 2019, Sudan. [12] | 0 | 42 | 0 | 0 | 18 | 0 | No |
| gB | FFPE | PCR | Sepahvand et al, 2019, Iran. [13] | 20 | 37 | 54.04 | 10 | 35 | 28.5 | Yes |
| Fresh | Real-time PCR | El Shazly et al, 2018, Egypt. [14] | 11 | 61 | 18 | 0 | 61 | 0 | Yes | |
| FFPE | ||||||||||
| FFPE | Real-time PCR | Shadood et al, 2018, Iraq [15] | 5 | 80 | 6.25 | 1 | 20 | 5 | No | |
| gB | FFPE | PCR | Bakhtiyrizade et al, 2017, Iran [16] | 0 | 150 | 0 | 2 | 150 | 1.3 | No |
| Real-time PCR | ||||||||||
| IE | FFPE | Real-time PCR | Mohammadizadeh et al, 2017, Iran [17] | 0 | 70 | 0 | 0 | 26 | 0 | No |
| gB | formlin- | Nested PCR | Karimi et al, 2016, Iran [18] | 26 | 50 | 58 | - | - | - | Yes |
| fixed | ||||||||||
| DNA | Fresh | Quantitative real | Mohammed et al, 2015, Iraq [19] | 9 | 50 | 23.7 | - | - | - | Yes |
| polymerase | time PCR | |||||||||
| pp65 | Fresh | quantitative PCR | Richardson et al, | 0 | 70 | 0 | 2 | 70 | 3 | No |
| 2015, New Zealand [20] | ||||||||||
| UL55 | Blood | PCR | Al.Baiati et al, 2014, Iraq [21] | 26 | 30 | 87 | 41 | 50 | 82 | Yes |
| UL97 | 20 | 30 | 67 | 33 | 50 | 66 | ||||
| gB | Fresh | PCR | Ahani et al, 2013, Iran [22] | 15 | 23 | 65.2 | - | - | - | Yes |
| UL122 and UL83 | paraffin | Real-time PCR | Barillas et al, | 2 | 27 | 7.4 | 0 | 20 | 0 | No |
| 2013, Mexican [23] | ||||||||||
| Fresh | Real-time PCR | Antonsson et al, 2012, Australia [24] | 0 | 54 | 0 | - | - | - | No | |
| Paraffin | PCR | Eghbali et al, 2012, Iran [25] | 2 | 24 | 8.3 | 0 | 24 | 0 | No | |
| gB | FFPE | Nested-PCR | Harkins et al, 2010, Birmingham [26] | 4 | 8 | 50 | 1 | 4 | 25 | Yes |
| IE2 | Fresh | PCR | Tsai et al, 2005, Taiwan [27] | 47 | 62 | 76 | 8 | 12 | 67 | Yes |
| Ig | Test | Studies | Cases | Controls | Significant Correlation | ||||
| No. | No. | Positivity | No. | No. | Positivity | ||||
| Positive | Tested | Positive | Tested | ||||||
| IgG | ELISA | Salman et al, 2020, Iraq. [28] | 67 | 71 | 94.3 | 19 | 20 | 95 | Yes |
| IgG | Indirect chemi-luminescent immunoassay | Surendran et al, 2019, India. [29] | 88 | 88 | 100 | 42 | 42 | 100 | No |
| IgM | 0 | 88 | 0 | 0 | 42 | 0 | |||
| IgG | ELISA | Al Nuimi et al, 2018, Iraq. [30] | 60 | 60 | 100 | 9 | 10 | 90 | Yes |
| IgM | 5 | 60 | 8.3 | 0 | 10 | 0 | |||
| IgG | ELISA | El Shazly et al, 2018, Egypt. [13] | 40 | 40 | 100 | 41 | 41 | 100 | Yes |
| IgM | 0 | 40 | 0 | 0 | 41 | 0 | |||
| IgG | ELISA | Mohammed et al, 2015, Iraq. [31] | 30 | 30 | 100 | 30 | 30 | 100 | Yes |
| IgM | 23 | 30 | 76.7 | 15 | 30 | 50 | |||
| IgG | Standard enzyme immunoassays | Richardson et al, 2015, New Zealand. [20] | 49 | 70 | 70 | - | - | - | Yes |
| IgG | ELISA | Al-Saady et al, 2014, Iraq. [32] | 30 | 30 | 100 | 91 | 98 | 93 | Yes |
| IgM | 18 | 30 | 60 | 12 | 98 | 12 | |||
| IgG | Enzyme immunoassays | Richardson et al, 2004, Australia. [33] | 122 | 208 | 59 | 97 | 169 | 57 | Yes |
| Ag | Test | Studies | Cases | Controls | Significant Correlation | ||||
| No. | No. | Positivity | No. | No. | Positivity | ||||
| Positive | Tested | Positive | Tested | ||||||
| IE | Immunohistochemistry | Costa et al,2019, Norway. [34] | 55 | 75 | 73 | 2 | 26 | 8 | Yes |
| Early Ag | Immunohistochemistry | Al Nuimi et al, 2018, Iraq. [35] | 56 | 60 | 93 | 2 | 10 | 20 | Yes |
| Late Ag | 37 | 60 | 61.7 | 7 | 10 | 70 | |||
| IE | Immunohistochemistry | Cui et al,2018, china. [36] | 62 | 68 | 92.6 | 70 | 146 | 47.9 | Yes |
| LA | 62 | 68 | 92.6 | 78 | 146 | 53.4 | |||
| E/IE | Immunohistochemistry | El Shazly et al, 2018, Egypt. [13] | 21 | 61 | 34 | 0 | 61 | 0 | Yes |
| PP65 | 49 | 61 | 80 | 15 | 61 | 25 | |||
| IE | immunohistochemistry | Mohammadizadeh et al, 2017, Iran. [16] | 0 | 70 | 0 | 0 | 26 | 0 | No |
| IE | Immunohistochemistry | Rahbar et al, 2017, Norway. [37] | 62 | 62 | 100 | 42 | 42 | 100 | Yes |
| L | 46 | 62 | 75 | 28 | 42 | 68 | |||
| Late Ag | Immunohistochemistry | Ahmed et al, 2016, Egypt. [38] | 47 | 107 | 43.9 | - | - | - | Yes |
| IE1 | Immunohistochemistry | Mohammed et al, 2015, Iraq. [31] | 34 | 38 | 89.5 | 3 | 30 | 10 | Yes |
| L | 35 | 38 | 92.1 | 0 | 30 | 0 | |||
| IE1/2 | immunohistochemistry | Harkins et al, 2010, Birmingham. [26] | 38 | 39 | 97 | 24 | 38 | 63 | Yes |
| Ultra- sensitive version of chromogenic in situ hybridization | Alajeely et al, 2019, Iraq. [39] | 8 | 20 | 40 | 0 | 20 | 0 | Yes | |
| Ultra- sensitive version of chromogenic in situ hybridization | Ali et al,2018, Iraq. [40] | 17 | 30 | 56.7 | 0 | 15 | 0 | Yes | |
| In situ hybridization | Rahbar et al,2017, Norway. [37] | 23 | 23 | 100 | - | - | - | Yes | |
| IE1 | In situ hybridization | Harkins et al,2010, Birmingham. [26] | 16 | 18 | 89 | 11 | 18 | 61 | Yes |
Detection of CMV Infection by PCR
Out of 16 studies, 7 studies used PCR method, 7 studies used Real-time method and 2 study used Nested PCR method. Different samples including fresh tissue, FFPE, and blood were evaluated by PCR methods for the presence of virus genome. In all studies that observed the presence of CMV genome in the samples, the prevalence of CMV genome in cancer samples was higher than normal samples. Virus detection was reported from 0 to 87% in cancer samples and from 0 to 82% in normal samples. The highest rate of virus detection in cancer samples was reported in the study of Al.Baiati et al, In Iraq and the lowest prevalence of the virus was reported in Sudan, Australia, Iran and New Zealand. Most of the studies related to PCR method are related to Iran, so that out of 16 studies, six studies have been done in Iran.
Serology of CMV Infection for Breast Cancer
Out of a total of studies, serological methods have been used in 8 studies. Of all serology studies, 8 studies examined anti-CMV IgG, 5 studies examined anti-CMV IgM, and 5 studies examined both antibodies. The level of anti-CMV IgG was higher than anti-CMV IgM in all studies and also the level of antibodies was higher in cancer samples than normal samples. The prevalence range of anti-CMV IgG was 70 to 100% and anti-CMV IgM was 0 to 76.7% in cancer samples and 90 to 100% for anti-CMV IgG and 0 to 57% for anti-CMV IgM in normal samples. The highest prevalence of anti-CMV IgG was from Iraq, Egypt and India and for anti-CMV IgM were from Iraq and Australia. The lowest level of anti-CMV IgG was observed in studies related to New Zealand and India and for anti-CMV IgM was observed in studies related to Egypt and India.
Detection of CMV Infection by IHC and ISH
The use of methods other than PCR, Real-time PCR, and ELISA that included IHC and ISH accounted for 13 of the total studies. There were fewer studies using ISH method than the other methods, so that among the total studies, in 4 studies ISH method and 9 studies IHC method was used. The results of using these methods showed a higher prevalence of virus in cancer samples than normal samples. Detection of virus antigens employing IHC in cancer and normal samples was reported to be 0 to 100%, while the rate of ISH was 40 to 100% and 0 to 67%, respectively. The highest prevalence of the virus was reported in Norway using IHC and ISH. These two techniques are widely used in different studies, especially from conducted investigation in Iraq.
Discussion
In this systematic review, we tried for the first time to examine the prevalence of HCMV in breast cancer. Among the 29 studies that were finally reviewed, the prevalence of CMV in breast cancer samples in studies that used PCR methods was between 0 and 87% (Table 1). In serological studies the seroprevalence of anti-CMV IgG and IgM was between 70-100% and 0-76.7% respectively (Table 2), and in other studies that served methods like IHC and ISH, the prevalence of the CMV was reported to be 0-100% and 40-100%, respectively (Table 3). In all studies that examined the prevalence of CMV in both cancer and healthy groups, the results indicated that the virus had higher frequency in tumor versus normal samples (Table 1-3).
Due to the advantages of PCR-based methods including conventional PCR. Real-time PCR, and Nested-PCR they were the most widely used methods in these studies, so that 16 studies from a total of studies served these methods (Table 1). The advantages of these methods include the sensitivity and rapidity of virus antigens detection than other methods, applicable for archived samples, the ability to detection virus antigens and its genome in various samples such as whole blood, leukocytes, plasma, body fluids (urine, CSF, BAL) or any other tissue (tissue biopsy samples) [41-42]. Although the Real-time PCR is more expensive than the antigen detection methods, it is faster and more automated and provides continuous monitoring of patients who are at risk of CMV for prophylactic treatment and determination of response to treatment [43]. Lack of distinction between active and inactive infections is one of the most important limitations of PCR-based methods [44], which causes other methods such as serology to be used in some of studies, as almost one third of the studies in this research have used serological methods such as ELISA (Table 2). Advantages of ELISA comprising of simplicity, cheapness and ability of determining the history of infection (early or late) make this method suitable for evaluating the presence of virus [42]. However, the less sensitivity and higher false results convinced some researchers to use other methods such as IHC or ISH to assess the presence of the virus [45]. Of the 29 studies reviewed, 13 studies used IHC or ISH techniques (Table 3). Since techniques such as IHC is the gold standard for the identification of CMV antigens presence in fresh tissue and FFPE, some of our studies served these techniques to accurately investigate the presence of viral antigens in malignant epithelial cells or non-epithelial cells [46].
Investigations have shown that all viral antigens are not required for tumorigenesis. CMV immediately early antigen (IE Ag) is known to be necessary for regulating the expression of other viral genes, replication, and cell transformation, therefore, in most studies, IE Ag has been evaluated as a target antigen by IHC [47-48]. In addition, this Ag, along with glycoprotein B or UL55, were the most widely used genes in evaluation of CMV genome presence by PCR methods (Table 1, 3). Glycoprotein B is considered as a virus ligand and essential for infectivity of virus [49]. Glycoprotein B nucleotide sequence is highly conserved, explaining its utilization as a detection target in various studies [49]. Although these studies have evaluated the virus in a variety of tissues, including fresh, FFPE, and blood, the major part of virus detection using this gene was reported in blood and fresh tissue [21-22]. In serological studies, among the all anti-CMV antibodies, the IgG and IgM were evaluated as a major indicator for viral presence (Table 2). The prevalence of anti-CMV IgG was examined in all serological studies, but the prevalence of anti-CMV IgM was examined only in three-quarters of studies along with anti-CMV IgG (Table 2). The results of studies showed a very high prevalence of anti-CMV IgG in both cancer and normal groups compared to the serum prevalence of anti-CMV IgM (Table 2). Due to the higher prevalence of anti-CMV IgG and especially anti-CMV IgM in the cancer group than the normal group (Table 2), serological studies suggest the probable role of the CMV in the development of breast cancer (Table 2). Since anti-CMV IgM can be present in both recent infection and viral reactivation which shows non-specificity for primary infection, the possibility of false positive results, and its inefficiency in the cases of immunocompromised patients, is not proposed as a highly useful indicator of virus infection [42]. On the other hand, anti-CMV IgG, which indicate previous exposure with the virus, can considered as a good indicator for last presence of HCMV infection [42].
Among serological studies, the highest prevalence of anti-CMV IgG has been reported in Asian countries such as Iraq, India, and African countries such as Egypt, as well as the highest prevalence of anti-CMV IgM was related to Iraq (Table 2). The highest viral detection rate based on PCR techniques was related to Asian countries such as Iraq, Iran (Zabul) and Taiwan (Table 1). The highest prevalence of the virus detection in tissue samples by means of IHC and hybridization methods was reported from Asian countries such as Iraq and China and European countries including Norway and the Birmingham, as well (Table 3). In general, the results obtained from different methods illustrate the high prevalence of CMV in Asian and African countries and its relatively lower prevalence in European and South American countries (Table 1-3). Since the prevalence of this virus directly depends on environmental factors such as social and economic conditions and even the cultural traditions, many studies have reported that low income and social status in third world countries is related to the higher prevalence of CMV [50-52]. For example, in areas such as Iraq, where families have a population of more than three, the prevalence of anti-CMV IgG and IgM determined to have an increasing trend [50].
The comparative analysis demonstrates the studies which employed serological and IHC methods identified higher presence of infection than PCR methods (Table 1-3). In this line it should be mentioned that sometimes even in CMV-positive tumor samples, many tumor cells do not contain the virus genome. This may be explained by specific event during the carcinogenesis. The findings suggest the role of two mechanisms of hit-run and oncomodulation. Oncomodulation defines as the mechanism of enhancing cellular malignancy following CMV infection [53]. This mechanism is carried out through specific CMV proteins including IE, US28, pp65, and ul44 which disrupts signaling pathways, transcription factors and tumor suppressor proteins in the epithelium of tumors and participates in increasing mutations, angiogenesis and facilitates the immune system evasion for tumor progression [11]. The ‘hit and run’ hypothesis is suggested as a main mechanism involved in viral transformation. In “hit-and-run” hypothesis, the virus can exert its long-term effect on cellular process to drive transformation when viral genomes are not present, supporting the importance of last exposure with viral infections [54].
In the case of anti-CMV therapies, as a result of virus ability in establishing latency in infected cells, no definitive treatment has been proposed for the infection [10]. However, the findings of previous studies in CMV- associated gliomablastoma provided promising results that have been achieved by targeting CMV in these patients using the therapies including the drug of valganciclovir, DC vaccine, and anti-HCMV targeted T cell therapy [10]. Some investigations introduce the utilization of a combination of therapies that induce the expression of viral lytic phase genes and expose the virus-containing tumor cells, is the best treatment strategy for herpesviruses in their latency form [10].
In conclusion, collectively, the majority of studies highlighted the possible role of CMV in developing breast carcinoma. Reviewing of all data showed the prevalence rate of CMV in patients with breast cancer is higher in low-income and third world countries than developed countries, proposing CMV prevalence may be affected by socio-economic status and environmental factors. The survey of methodology represents each technique has different sensitivity, specificity, and reliability in comparison with others. The characteristic of CMV in establishing latent form of infection is a significant ongoing limitation for anti-CMV therapies and the future investigation should focus on finding a promising solution for this issue.
Acknowledgments
We are thankful to Akram Valizadeh for support the current study.
Conflict of Interests
All authors declared that they have no conflict of interest.
Funding/Support
No.
Financial Disclosures
The authors have no financial interest related to the material in the manuscript.
References
- Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle, Siegel Rebecca L., Torre Lindsey A., Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2018;68(6). CrossRef
- Subtypes of benign breast disease as a risk factor of breast cancer: A systematic review and meta analyses Salamat F, Niakan B, Keshtkar A, Rafiei E, Zendehdel M. Iranian journal of medical sciences.2018;43(4):355.
- Use of Statistics to Assess the Global Burden of Breast Cancer Parkin DM, Fernandez LMG. The Breast Journal.2006;12(s1). CrossRef
- Breast cancer statistics, 2013 DeSantis Carol, Ma Jiemin, Bryan Leah, Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2013;64(1). CrossRef
- Cancer incidence and mortality worldwide: IARC CancerBase Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al . Globocan.2012;2013(1):11.
- The global health burden of infection-associated cancers in the year 2002 Parkin Donald Maxwell. International Journal of Cancer.2006;118(12). CrossRef
- Viral infections and breast cancer – A current perspective Gannon O.M., Antonsson A., Bennett I.C., Saunders N.A.. Cancer Letters.2018;420. CrossRef
- A Review of the Potential Role of Human Cytomegalovirus (HCMV) Infections in Breast Cancer Carcinogenesis and Abnormal Immunity Geisler Jürgen, Touma Joel, Rahbar Afsar, Söderberg-Nauclér Cecilia, Vetvik Katja. Cancers.2019;11(12). CrossRef
- The Story of Human Cytomegalovirus and Cancer: Increasing Evidence and Open Questions Michaelis Martin, Doerr Hans W., Cinatl Jindrich. Neoplasia.2009;11(1). CrossRef
- The emerging role of human cytomegalovirus infection in human carcinogenesis: a review of current evidence and potential therapeutic implications Nauclér Cecilia Söderberg, Geisler Jürgen, Vetvik Katja. Oncotarget.2019;10(42). CrossRef
- The role of viruses in adenocarcinoma development Nakhaie Mohsen, Charostad Javad, Kaydani Gholam Abbas, Faghihloo Ebrahim. Infection, Genetics and Evolution.2020;86. CrossRef
- Molecular Detection of Epstein-Barr Virus and Human Cytomegalovirus Antigen Expression in Breast Cancer in Khartoum State, Sudan 2018.2019 Aidrous RE, El Hussein ARM, Dahawi SH, Elkhidir IM, Enan KA. .
- Human Cytomegalovirus DNA among Women with Breast Cancer Sepahvand Peyman, Makvandi Manoochehr, Samarbafzadeh Alireza, Talaei-Zadeh Abdulhasan, Ranjbari Nastaran, Nisi Nilofar, Azaran Azarakhsh, Jalilian Shahram, Pirmoradi Roya, Makvandi Kimia, Ahmadi Angali Kambiz. Asian Pacific Journal of Cancer Prevention.2019;20(8). CrossRef
- Detection of Human Cytomegalovirus in Malignant and Benign Breast Tumors in Egyptian Women El Shazly Dalia F., Bahnassey Abeer A., Omar Omar S., Elsayed Elsayed T., Al-Hindawi Ali, El-Desouky Eman, Youssef Hend, Zekri Abdel-Rahman N.. Clinical Breast Cancer.2018;18(4). CrossRef
- Correlation of Breast Cancer with the Epstein Bar Virus and Human Cytomegalovirus Frequency and the Expression of Estrogen Receptor-Beta and IL-6 Receptor in Iraqi Women A. L. Shadood Hayder Kadhim, Atiya Saad Abdul-Aziz, Kardar Gholam Ali. Natural Science.2018;10(05). CrossRef
- Almost Complete Lack of Human Cytomegalovirus and Human papillomaviruses Genome in Benign and Malignant Breast Lesions in Shiraz, Southwest of Iran Bakhtiarizadeh Sahar, Hosseini Seyed Younes, Yaghobi Ramin, Safaei Aliakbar, Sarvari Jamal. Asian Pacific Journal of Cancer Prevention.2017;18(12). CrossRef
- Evaluation of human cytomegalovirus antigen expression in invasive breast carcinoma in a population of Iranian patients Mohammadizadeh Fereshteh, Mahmudi Fatemeh. Infectious Agents and Cancer.2017;12(1). CrossRef
- Relative Frequency of Cytomegalovirus (CMV) in Tissue Samples of Women with Breast Cancer in Sanandaj, Iran. Karimi Maryam, Hosseini* Seyedeh Zeinab, Nikkhoo Bahram, Mohammadi Farid Soleimani-. International Journal of Bioassays.2016;5(03). CrossRef
- Investigation the role of human cytomegalovirus in the invasive ductal breast carcinoma using real time PCR Mohammed AH, Kadhim HS, Hussein AA. Int J Curr Microbiol App Sci.2015;4(1):537-542.
- Cytomegalovirus and Epstein-Barr Virus in Breast Cancer Richardson Ann K., Currie Margaret J., Robinson Bridget A., Morrin Helen, Phung Yen, Pearson John F., Anderson Trevor P., Potter John D., Walker Logan C.. PLOS ONE.2015;10(2). CrossRef
- Molecular Detection of Human Cytomegalovirus genes in infertile and breast cancer women in Baghdad province Hussein AMAB , Rebah N, Jabbar2 , Mohammed A.K, Al-Saady3 , A.Muhsin3 aM . Int J Curr Microbiol App Sci.2014;3(4):316-322.
- Detection of Human Cytomegalovirus in Breast Cancer by Polymerase Chain Reaction (PCR) Ahani N, Alipour M, Nikravesh A. Shahid Beheshti University Of Medical Of Sciences.2013;:8.
- Is human cytomegalovirus associated with breast cancer progression? Utrera-Barillas Dolores, Valdez-Salazar Hilda-Alicia, Gómez-Rangel David, Alvarado-Cabrero Isabel, Aguilera Penélope, Gómez-Delgado Alejandro, Ruiz-Tachiquin Martha-Eugenia. Infectious Agents and Cancer.2013;8(1). CrossRef
- Exploring the Prevalence of Ten Polyomaviruses and Two Herpes Viruses in Breast Cancer Antonsson Annika, Bialasiewicz Seweryn, Rockett Rebecca J., Jacob Kevin, Bennett Ian C., Sloots Theo P.. PLoS ONE.2012;7(8). CrossRef
- Frequency of cytomegalovirus (CMV) in benign and malignant tumors Eghbali M, Ghane M, Mirinargesi M. International Journal of Molecular and Clinical Microbiology.2012;2(2):175-179.
- Detection of human cytomegalovirus in normal and neoplastic breast epithelium Harkins Lualhati E, Matlaf Lisa A, Soroceanu Liliana, Klemm Katrin, Britt William J, Wang Wenquan, Bland Kirby I, Cobbs Charles S. Herpesviridae.2010;1(1). CrossRef
- Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues Tsai Ju-Hsin, Tsai Chung-Hung, Cheng Min-Hsiung, Lin Shyh-Jye, Xu Fang-Ling, Yang Chi-Chiang. Journal of Medical Virology.2004;75(2). CrossRef
- Seroprevalence of human cytomegalovirus in iraqi breast cancer patients Salman OH, Al-Azzawi RH. Plant Archives.2020;20:729-731.
- Breast Cancer Association with Cytomegalo Virus—A Tertiary Center Case-Control Study Surendran Anilkumar, Chisthi Meer M.. Journal of Investigative Surgery.2017;32(2). CrossRef
- Serodiagnosis of Human Cytomegalovirus in Iraqi Breast cancer and fibroadenoma patients AlNuaimi BN, Al-Azzawi RH, Naji RZ. Current Research in Microbiology and Biotechnology.2018;6(1):1466-1469.
- Investigation the role of human cytomegalovirus in the invasive ductal breast carcinoma Mohammed AhmedHasan, Kadhim HaiderSabah, Ghani AlaaHussein. Clinical Cancer Investigation Journal.2015;4(2). CrossRef
- Serodiagnosis of Human Cytomegalovirus in infertile and breast cancer women in Baghdad province Al-Saady M. Int J Curr Microbiol App Sci.2014;3(4):290-295.
- Cytomegalovirus, Epstein–Barr virus and risk of breast cancer before age 40 years: a case–control study Richardson A K, Cox B, McCredie M R E, Dite G S, Chang J-H, Gertig D M, Southey M C, Giles G G, Hopper J L. British Journal of Cancer.2004;90(11). CrossRef
- Human cytomegalovirus infection is correlated with enhanced cyclooxygenase-2 and 5-lipoxygenase protein expression in breast cancer Costa Helena, Touma Joel, Davoudi Belghis, Benard Melinda, Sauer Torill, Geisler Jürgen, Vetvik Katja, Rahbar Afsar, Söderberg-Naucler Cecilia. Journal of Cancer Research and Clinical Oncology.2019;145(8). CrossRef
- Association of human cytomegalovirus with HER2 proto-oncogene overexpression in iraqi breast cancer patients. Biochem Al-Nuaimi BN, Al-Azzawi RH, Naji RZ. Cell. Arch.2019;19(1):1691-1698.
- Protein and DNA evidences of HCMV infection in primary breast cancer tissues and metastatic sentinel lymph nodes Cui Jian, Wang Qian, Wang Hai-Bo, Wang Bin, Li Ling. Cancer Biomarkers.2018;21(4). CrossRef
- Low Expression of Estrogen Receptor-α and Progesterone Receptor in Human Breast Cancer Tissues Is Associated With High-Grade Human Cytomegalovirus Protein Expression Rahbar Afsar, Touma Joel, Costa Helena, Davoudi Belghis, Bukholm Ida Rashid, Sauer Torill, Vetvik Katja, Geisler Jürgen, Söderberg-Naucler Cecilia. Clinical Breast Cancer.2017;17(7). CrossRef
- Immunohistochemical detection of human cytomegalovirus, Epstein-Barr virus and human papillomavirus in invasive breast carcinoma in Egyptian women: A tissue microarray study Ahmed Rehab Allah, Yussif Shaimaa M.. Journal of Solid Tumors.2016;6(2). CrossRef
- Assessment of breast cancer tissue microenvironment for human cytomegalovirus infection and CD4- and CD8- positive tumor infiltrating lymphocytes Alajeely AAA, Mahmood AS, Zangor JR, Ali SHM . Biochem Cell Arch.2019;19:2499-2505.
- Impact of human cytomegalovirus infection associated with the expressed protein of mutated BRCA1 gene in breast tissues from a group of Iraqi female patients with breast carcinoma Ali SHM, Almahbobi TF, Al-Bayaa YJ, Al-Alwany SHM. Research Journal of Pharmacy and Technology.2018;11(4):1505-1512.
- Polymerase Chain Reaction Garibyan Lilit, Avashia Nidhi. Journal of Investigative Dermatology.2013;133(3). CrossRef
- Overview of the Diagnosis of Cytomegalovirus Infection A. Ross S., Novak Z., Pati S., B. Boppana S.. Infectious Disorders - Drug Targets.2011;11(5). CrossRef
- Cytomegalovirus infection in transplant recipients Azevedo LS, Pierrotti LC, Abdala E, Costa SF, Strabelli TM, Campos SV, Ramos JF, Latif AZ, Litvinov N, Maluf NZ, Caiaffa Filho HH, Pannuti CS, Lopes MH, Santos VA, Linardi CC, Yasuda MA, Marques HH. Clinics.2015;70(7). CrossRef
- Application of PCR-Based Methods To Assess the Infectivity of Enteric Viruses in Environmental Samples Rodríguez Roberto A., Pepper Ian L., Gerba Charles P.. Applied and Environmental Microbiology.2008;75(2). CrossRef
- Serological and Molecular Comparison Study for Diagnosis of Cytomegalovirus Infection in aborted Pregnant Women in Iraq Al-Taie Anmar, Abdullah Basim, Al-Attar Mozahim. Rafidain Journal of Science.2018;27(4). CrossRef
- A Comparison of CMV Detection in Gastrointestinal Mucosal Biopsies Using Immunohistochemistry and PCR Performed on Formalin-fixed, Paraffin-embedded Tissue Mills Anne M., Guo Frances P., Copland Andrew P., Pai Reetesh K., Pinsky Benjamin A.. American Journal of Surgical Pathology.2013;37(7). CrossRef
- Immediate-Early (IE) gene regulation of cytomegalovirus: IE1- and pp71-mediated viral strategies against cellular defenses Torres Lilith, Tang Qiyi. Virologica Sinica.2014;29(6). CrossRef
- Immediate–early viral gene regulation and function. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. 2007 Stinski MF, Meier JL. .
- Diagnostic Consequences of Cytomegalovirus Glycoprotein B Polymorphisms Novak Z., Chowdhury N., Ross S. A., Pati S. K., Fowler K., Boppana S. B.. Journal of Clinical Microbiology.2011;49(8). CrossRef
- Cytomegalovirus seroprevalence in women with bad obstetric history in Kirkuk, Iraq Aljumaili Zainab Khalil Mohamed, Alsamarai Abdulghani Mohamed, Najem Wesam Suhail. Journal of Infection and Public Health.2014;7(4). CrossRef
- Socioeconomic disparities in the seroprevalence of cytomegalovirus infection in the US population: NHANES III DOWD J. B., AIELLO A. E., ALLEY D. E.. Epidemiology and Infection.2008;137(1). CrossRef
- Congenital cytomegalovirus infection is associated with high maternal socio-economic status and corresponding low maternal cytomegalovirus seropositivity Basha James, Iwasenko Jenna M, Robertson Peter, Craig Maria E, Rawlinson William D. Journal of Paediatrics and Child Health.2014;50(5). CrossRef
- Oncomodulation by human cytomegalovirus: evidence becomes stronger Michaelis Martin, Doerr Hans Wilhelm, Cinatl Jindrich. Medical Microbiology and Immunology.2009;198(2). CrossRef
- Viral hit and run-oncogenesis: Genetic and epigenetic scenarios Niller Hans Helmut, Wolf Hans, Minarovits Janos. Cancer Letters.2011;305(2). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details