High Prevalence of Mitochondrial tRNA A3243G Mutation in Invasive Breast Cancer
Download
Abstract
Background & Aim: Mitochondria play vital roles in various cellular activities, such as energy production, maintaining the redox balance of the cell, the regulation of cellular proliferation and differentiation, and programmed cell death. Mitochondrial tRNA mutations are associated with many pathological conditions and numerous diseases have been associated with them. Therefore, the objectives of this study were to evaluate the presence and the frequency of mutations in the selected tRNAs in breast cancer patients and to correlate these mutations with the clinicopathological characters.
Materials & Methods: The is a cross sectional study, where seventy seven breast tumors and adjacent non-tumorous (normal) tissue samples were analyzed by PCR- RFLP, and direct DNA sequencing. In this study three mt-tRNAs wre selected, two of them are the most important disease associated mt-tRNAs i.e tRNALue (UUR), tRNALys and one of the least pathogenic associated tRNA which is tRNAarg. Statistical analysis was used to determine the relationship between the detected mutations and the clinicopathologhical characters.
Results: The tRNAleu (UUR) A3243G mutation occurring at the highly conserved location was detected in 4 samples (5.2%) of both breast cancer and adjacent non-tumorous tissue except one detected only in tumor tissue. Another A8343G mutation was detected in tRNAlys. There was no significant association between these mutations and clinicopathological parameters (P. value > 0.05).
Conclusion: The detection of high frequent A3243G mutations in patients with invasive breast cancer suggests role of this mutation in the carcinogenesis. This mutation affects complex I, reduces translation of its core subunits, which can contribute to the impairment of oxidative phosphorylation and increase ROS production, leading to cancer progression.
Introduction
Breast is the most frequently detected cancer in most countries in the world (154 of 185 countries) and also it is the leading cause of cancer mortality in more than 100 countries [1]. According to Globocan report 2018, there is more than 2 million newly diagnosed cases of female breast cancer representing about 1 in 4 of all cancer cases in women [1]. The rate of female breast cancer incidence differ among countries from 19.9/ 100000 women in Eastern countries of Africa to 89.9 /100000 women in Western Europe. The age standardized incidence rate for breast cancer in India is 22.9 per 100000 and it is the most common cancer among Indian females preceded by cervical cancer, with an estimated 115,251 new diagnosed cases and 53,592 breast cancer deaths [2].
Mitochondria are distinctive organelles in eukaryotic cells which can play important roles in both cellular life and death. Mitochondria play vital roles in various cellular activities, such as steroid and heme synthesis, regulation of intracellular calcium ions, balancing the redox process in the cell, many important metabolic pathways such as oxidation of fatty acid, the regulation of several cellular activities like proliferation and differentiation, and programmed cell death which are important key features to the development of malignant transformation [3-4].
Despite that, mitochondria are the only organelle of an animal cell which has its own genome, a great number (about 1500) of nuclear encoded gene products are crucial for the success performance of normal mitochondrial function and biogenesis. Mitochondrial DNA molecule is small, compact closed-circular double-stranded DNA, multiple in copy. Mitochondrial genome is composed of 16,569 bp and encodes for 37 genes which include 13 polypeptides, 2 rRNA and 22 tRNA genes [5]. The 22 mitochondrial tRNAs represent the least requirement for the translation of mitochondrial genes which encode the thirteen polypeptides [6].
Most tRNAs from all living organisms have a highly conserved cloverleaf structure involving four stems and three loops (Figure 1).
Figure 1. Mitochondrial Classical tRNA with Normal Cloverleaf Secondary Structure (left) and the two Unusual mt-tRNA (right) with Loss of Complete D-arm, the Case of tRNASer (AGY), and the Case of tRNASer (UCN) which has a Shortened Connector (single base) and an Extended Anticodon Stem (arrows shows the place of unusual features).
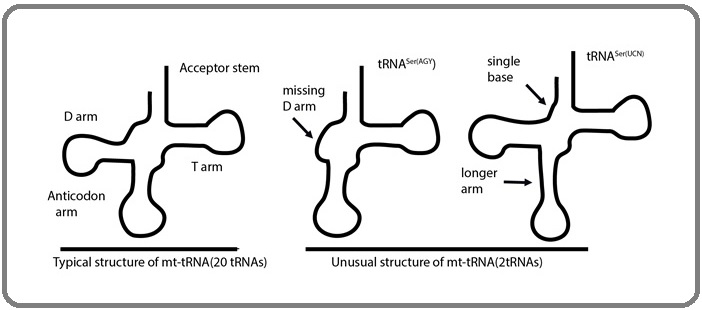
Compared to cytoplasmic tRNAs, mammalian mitochondrial tRNAs in general have some different properties and many of them have unusual structure, they are less conserved with the variation in the level of conservation within tRNA families, D and T arms can vary to a large extent with a tendency to have small loops and they are more variable in size than cytoplasmic tRNAs [7]. Although some deviations in stem and loop sizes of mitochondrial tRNA, only two mt-tRNAs have a typical structure, mt-tRNASer (AGY) has no D-arm, while mt-tRNASer (UCN) has a single nucleotide connecting the D and anticodon stems (Figure 1).
The association between mitochondrial respiratory change and cancer was initially introduced by Otto H. Warburg [8]. Later on, after the discovery of human mitochondrial genome and its sequences, it was extensively investigated in all human cancer and mutations in this small mitochondrial DNA had been identified in all types of malignancies [9]. The first pathogenic point mutation in the mitochondrial DNA (mtDNA) was characterized in 1988 [10]. After that, plenty of mitochondrial DNA mutations have been connected to various human diseases and about half of these mutations are located within tRNA genes, which is striking given that all mt-tRNAs gene sequences cover no more than 10% of the whole mitochondrial genome [11].
The large portion of mitochondrial DNA pathogenic conditions are particularly associated with mutations in the 22 mitochondrial tRNAs, but there are few studies investigating mitochondrial tRNA mutations in cancers. In the present study we selected three mt-tRNAs, two of them are the most important disease associated mt-tRNAs i.e tRNALue(UUR), tRNALys and one of the least pathogenic associated tRNA which is tRNAarg. The purpose of this selection is to compare the prevalence of mutation in these tRNAs in cancer. The objectives of this study were to evaluate the presence and the frequency of mutations in the selected tRNAs in breast cancer patients from the Indian population, and to correlate these mutations with the clinicopathological characters of BC.
Materials and Methods
Sample preparation
The present study protocol was approved by the Ethical Committee of Kamineni Hospital. A total of 100 paired archival paraffin blocks of breast cancer and adjacent normal breast tissue of the same patient were collected by the pathology department of Kamineni Hospital. All samples were taken after breast surgery and those blocks were included in the study where tumor tissues and adjacent normal were verified by an experienced pathologist. These BC samples were TNM staged as indicated by the American Joint Committee on Cancer. Sixty nine tumors were staged and eight were only described as Invasive Ductal Carcinoma (IDC), without additional information.
DNA isolation
DNA was extracted from consecutive sections obtained from formalin fixed embedded paraffin blocks (FFEP) after identifying tumor and adjacent normal areas with histopathology as per the established method from our laboratory [12]. Due to difficulties in isolation of DNA from FFEP sufficient amount of DNA for the study was successfully isolated only from seventy seven pairs of breast cancer and its adjacent non-tumorous tissue, because of that only those samples included in the study.
PCR-RFLP
The target mitochondrial DNA fragments of 501bp, 200 bp and 810bp containing the selected tRNALeu, tRNA Lys, tRNA arg were amplified using polymerase chain reaction (PCR) technique, utilizing the selected primers given in Table 1.
| Genes | Forward/Reverse primers | Size of PCR products | Method |
| ND1+tRNA lue | TTGGATCAGGACATCCCGATGGTGCAG | ||
| GTTTTAGGGGCTCTTTGGTGAAGAGTT | 502bp | HaeIII | |
| Cox2+tRNA lys | 5'- TCG TCC TAG AAT TAA TTC CC -3 | ||
| 3’- GGG GGT AAT TAT GGT GGG CC -5 | 200 bp | AvaII | |
| ND3+tRNA Arg | CCAACCTCCTACTCCTCATTG | ||
| TGTTGTGTAGAGTTCAGGGGA | 810 bp | Sequencing |
PCR conditions were an initial denaturation at 94 oC for 4 minutes, then followed by series of 35 cycles of denaturation at 94 oC for 30 sec, 62 oC for 45 seconds and 72 oC for 45 seconds. The final extension was 72 oC for 5 minutes. PCR amplification was done in a 50 µl reaction volume containing 2 mM of each dNTP, 50mM KCl, 1.5 mM MgCl2, 10mM Tris-HCl (PH 8.3), 10 pmol of each primer, and 1.5 U of Taq DNA polymerase (cat #105917 BangaloreGeni, India). All PCR amplicons were checked on 2 % agarose gel electrophoresis in 1XTBE buffer utilizing a 50 bp DNA ladder as a marker for evaluating the size of the amplicons. HaeIII restriction enzyme was used to screen for A3243G mutations in tRNA Leu(UUR) and AvaII was used to screen for A8343G mutation in tRNAlys. Then it was loaded on 12 % Polyacrylamide Gel Electrophoresis (PAGE) and electrophoresed in 1X TBE buffer under 200 V for 2 hours after electrophoresis the gel was ethidium bromide stained for 10 minutes.
DNA sequencing
The mtDNA fragment containing the tRNAArg gene was sequenced commercially in all breast cancer samples at Vimta lab. Hyderabad.
Statistical Analyses
The strength of the relationship between each categorical variable was assessed by χ2-test. All statistical analysis was carried out with STATA/MP 13 software. A p-value less than 0.05 was considered statistically significant.
Results
Seventy seven paired samples of breast cancer and its adjacent non-tumorous tissue were analyzed in this study for the presence of mutations in the three mt-tRNA regions amplified. The age range of the patients was 25 - 72 years. The patients were divided into two groups: one < 50 year and the other ≥ 50 years for evaluation (Table 2).
| Clinical pathological | No. of patients | P. Value | ||
| Characteristics | N=77 | |||
| No Mutation | Mutation | |||
| Age range/years | ||||
| <50 | 51 | 49 | 2 | 0.98 |
| ≥50 | 26 | 47 | 3 | |
| Histological type | ||||
| Invasive | 64 | 59 | 5 | 0.67 |
| In situ | 13 | 13 | 0 | |
| Stage | ||||
| I | 22 | 20 | 2 | 0.72 |
| II | 31 | 30 | 1 | |
| III | 11 | 10 | 1 | |
| IV | 5 | 5 | 0 | |
| No specific stage | 8 |
66.2% of BC patients were identified in the younger age group under the age of 50 year. The mean age of the patients was 48± 8 years. The breast tumors were classified as Invasive Ductal Carcinoma (IDC), Invasive Lobular Carcinoma (ILC) Ductal Carcinoma In Situ (DCIS). The tumors were also staged as I to IV based on the histopathological and clinical criteria. In this study, all cases were female breast cancer except one case of IDC male breast cancer (MBC) which was diagnosed at the age of 67 years.
The A3243G mt-DNA occuring in the tRNALeu (UUR), after PCR amplification gave a band of 502 bp which was RD with HaeIII enzyme, four DNA bands were observed in normal RD whereas only three bands were found in mutated DNA (Figure 2).
Figure 2. Showing Results of HaeIII Restriction Digestion of mtDNA 501 bp on 12% EtBr Stained PAGE, Four Bands Observed in Normal (A) and (B) whereas only three Bands with 266 bp Bands, (B) 3 and 4 Lane Indicating the Presence of Mutation and Loss of Restriction Site. The size of the DNA ladder (L) is 50bp from Invitrogen (Cat. Number: 10488099).
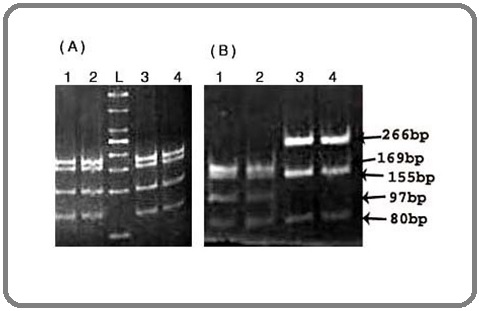
The mitochondrial tRNAleu A3243G transition mutation was detected in 4 samples (5.2%) of breast cancer and adjacent non-tumorous tissue. In 3 cases, both tumor and adjacent non-tumorous tissue had the mutation and in one case it was seen in tumor tissue alone. All mutations detected were homoplasmic. Mitochondrial tRNAlys is encoded by nucleotides 8295-8364 of the mitochondrial DNA heavy strand. The AvaII restriction enzyme was used to screen for the A8343G mutation. This mutation was found in one case of breast cancer tissue but not in its adjacent non-tumorous breast tissue. The mt-tRNA arginine gene was sequenced in all breast cancer DNA samples, but all of them showed no changed in DNA sequences.
Discussion
In the present study, three mt-tRNA genes i.e tRNAleu (UUR) and tRNAlys which are the most important mt-tRNAs with respect to pathogenicity were analyzed. Also in this study, mt-tRNAArg, which representing one of the least pathogenic associated tRNA was analyzed. Our results showed, that mt-tRNA mutations were present in BC from Indian patients. A total of five homoplasmic mutations were detected in the selected tRNAs for this study, among these mutations, four A3243G were detected in tRNAlueUUR three of them detected in both tumor and its adjacent normal breast tissue, whereas one mutation was found only in tumor but not in the adjacent normal tissue. Another homoplasmic A8343G mutation in tRNAlys was also found. The presence of these well-known pathogenic mutations in both tumor and adjacent normal tissue indicated that, they were germline mutations and were expected to have a role in the BC carcinogenesis. All detected mutations were found only in patients with invasive ductal carcinoma (IDC).
The uncovering of pathogenic point mutations in human mitochondrial genes that can cause neurodegenerative and neuromuscular disorders brings more scientific and clinical attention to mitochondrial genome, with particular interest in studying mt-tRNAs since most of the detected mitochondrial pathogenic mutations occur in these genes. Subsequently, the extensive studies on mitochondrial genome lead to rapidly increasing of mutation rates detected in mt-RNAs genes with only three mutations reported in 1990 and more than 230 in 2012 [13]. According to the latest data from MITOMAP (March 2015) about the frequency of mt-tRNA mutations, a total number of 252 mutations have been reported in all of the 22 mt-tRNAs. The maximum number of mutations was reported in tRNAleu (UUR) (33/13.1%) followed by tRNAlys (23/9.1%) and only 5 (2%) mutations were reported in tRNAarg.
It is interesting to note that, all human diseases which are directly associated with mutations in human cell tRNAs, these mutations have been found to exist only in mt-tRNAs. The main possible reason for that is a mutated mt-tRNA most probably not going to be compensated for by other tRNAs (as in nucleus there are multiple copies for each single tRNA) since each mitochondrial genome has a single copy of only the minimal necessary number (22 mt-tRNAs) and importing tRNA from the nucleus is rare in human [14-15].
The A3243G mutation is located at highly conserved genome location in the D loop of the mt-tRNALeu (UUR) (Figure 3), as a result of that A3243G has multiple consequence on structural stability, methylation, aminoacylation and codon recognition, leading to different cellular and metabolic alterations involving high glycolytic rate, more production of lactate, decreased glucose oxidation, lowered mitochondrial membrane potential, reduced ATP production, disturbed cellular calcium homeostasis with an increased cytosolic calcium load [16].
Figure 3. Conservational Sequence Alignment of Evolutionarily Diverse Organisms Reveals, that the Position of the m.3243 A >G Mutation in Mitochondrial tRNAleu (UUR) is Highly Conserved as Expected for Pathogenic Change.

It is worth mentioning that the overall functional and structural integrity of OXPHOS may be affected by mutations in mitochondrial tRNA genes, due to a general reduction in translation of mtDNA-encoded subunits mainly CI which is the most affected by these mutations, since half of its core subunits (7/14) are encoded by mtDNA. The association between mt-tRNA mutations with an increased risk of human cancer is not yet clear, but reported cases of the well known pathogenic A3243G mutation in colon cancer and renal cell carcinoma and the absence of this mutation from normal adjacent tissue suggest the involvement in the carcinogenesis process [17-18].
In this study the mean age for breast cancer patients was 48±8 years, which is similar to what has been reported from the Indian population [19], 19.5% cases (15 cases) from the present study were diagnosed with BC at a younger age, i.e. between 25-40 years, 49 cases (63.6%) were in the age group of 41-55 year and 13 cases (16.8%) were of age beyond 55 years (56-75years). These results indicated that most of the cases were diagnosed at an age below 55 years, which is less than the mean age of breast cancer patients in the US and European countries [20-23]. The mean age of patients who has mutations in tRNAs was found to be 46.6± 8.7 years which is even lower.
In this study, the statistical analysis of breast cancer clinicopathological characteristics revealed that, there was no significant association between patients’ age, and BC grade, stages and the presence of mitochondrial DNA mutations in mt-tRNAs. This is consistent with findings from previous studies showing a significant association between mtDNA variations and clinicopatholgogical parameters. For example, D-loop mutations were observed to be associated with poor prognosis in colon cancer as well as response to 5-Fluorouracil treatment [24]. In breast cancer, D-loop mutations, particularly in the highly polymorphic region known as D310 were associated with older age, estrogen, and progesterone receptor negative status, as well as poorer progression-free survival [25]. In a study conducted on breast cancer mtDNA mutations, histopathologic analysis showed a significant association between mitochondrial genome microsatellite instabilities and invasive lobular carcinoma (ILC) (74%) compared to IDC (19%) [26].
The A3243G mtDNA mutation in tRNAlys, is the most common mitochondrial disease associate point mutation that cause disease known as myopathy encephalopathy lactic acidosis and stroke-like episodes (MELAS), and also it can be linked with other diseases like maternally inherited type II diabetes, progressive external ophthalmoplegia (PEO) and deafness syndrome [27].
Mutations in mitochondrial tRNAs have been identified in different type of cancer, for example, Zhu et al 2005 revealed that mt mutations in BC can be identified in breast nipple aspirate fluid (NAF). Fifteen breast cancers, matched benign tissues and NAF were assessed and two mutations were found in tRNA isoleucine and another one in tRNA threonine[28]. Mitochondrial tRNAs mutation was also found in hepatocellular carcinoma, lung cancer and in oncocytic pituitary adenoma [29-31]. The functional consequences and the mechanism by which these mutations exert their influence in the carcinogenesis process need to be assessed and analyzed.
Pathology related mutations have been identified in all mt-tRNAs and there is a wide-ranging of variation in the outcome disease phenotypes, although understanding of how genotype relates to phenotype is not yet clear. Many of these differences can be clarified by variations in tissue segregation, heteroplasmy, the location of a mutation in mitochondrial tRNA and mutation threshold effect, but the exact molecular mechanisms by which mutations cause disease are somewhat poorly understood. The genotype–phenotype correlation of these mutations have been studied extensively, but no reliable indicators of how a mutation presents have yet been determined. Various possible molecular effects of mt-tRNAs mutations have been identified, including, affecting maturation of the primary transcript, the prevention of aminoacylation, transcription factor binding disruption, and difficulties in codon recognition with the most common feature for all mutated mt-tRNAs is loss of structure stability [32-33].
The findings of homoplasmic A3243G mutation, along with other mutations in evolutionary conserved site of mt-tRNA genes in tumor cells indicated that those cells may obtain their respiratory-deficient phenotype through mtDNA changes. The A3243G (MELAS) mutation is among the best well characterized mt-DNA defects and is commonly manifested as a severe disease. At the molecular level, this mutation is well-known to affect tRNALeu (UUR) aminoacylation and the wobble modification in the anticodon. Cybrid studies revealed that cells retain more than 95% of A3243G mutation in their mtDNA show acute respiratory deficient phenotype, a reduction in both oxygen consumption rates and electron transfer activities and display evidence of increased oxidative damage [16, 27].
In the present study, mt-tRNAarg showed no alteration in any of the analyzed breast cancer samples, which is similar to what have been published up to now since there are very limited numbers of pathogenic mutations describe in this RNA particularly. Some of the reported mt-tRNAarg mutations have been found associated with certain diseases like myopathy and progressive encephalopathy and only one mutation has been reported in endometrial tumor (T10364C) [32-33].
In conclusion, the detection of mitochondrial tRNAs mutations in BC, particularly (A3243G MELAS mutation), which is the most common one in relation to pathogenicity and arise in a highly conserved genome location and the detection of these mutations in patients with invasive BC only indicated that, these mutations may play a causative role in carcinogenesis. Also mitochondrial tRNAs mutations, particularly A3243G contribute to complex I (CI) dysfunction through an overall OXPHOS impairment which may allow cancer cells to progress toward malignant phenotype. Further studies, especially functional studies needed to confirm the role of these mutations in the carcinogenesis process.
Acknowledgements
This study was supported by Department of Science and Technology (DST) India. [Grant # (SR/SO/HS- 89/2004]. The authors would like to thank the members of Department of Pathology, Kamineni Hospital for providing the BC tissue samples.
Author Disclosure Statement
No competing financial interests exist.
References
- Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle, Siegel Rebecca L., Torre Lindsey A., Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2018;68(6). CrossRef
- Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008 Ferlay Jacques, Shin Hai-Rim, Bray Freddie, Forman David, Mathers Colin, Parkin Donald Maxwell. International Journal of Cancer.2010;127(12). CrossRef
- Mitochondria and cancer Wallace Douglas C.. Nature Reviews Cancer.2012;12(10). CrossRef
- The Role of Mitochondria in Apoptosis Wang Chunxin, Youle Richard J.. Annual Review of Genetics.2009;43(1). CrossRef
- Sequence and organization of the human mitochondrial genome Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H. L., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J. H., Staden R., Young I. G.. Nature.1981;290(5806). CrossRef
- The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA Helm M., Brule H., Degoul F., Cepanec C., Leroux J.-P., Giege R., Florentz C.. Nucleic Acids Research.1998;26(7). CrossRef
- Different pattern of codon recognition by mammalian mitochondrial tRNAs. Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., Young I. G.. Proceedings of the National Academy of Sciences.1980;77(6). CrossRef
- On the Origin of Cancer Cells Warburg O.. Science.1956;123(3191). CrossRef
- Somatic Mitochondrial DNA Mutations in Human Cancers Yu M. 2012;57:99-138. CrossRef
- Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies Holt I. J., Harding A. E., Morgan-Hughes J. A.. Nature.1988;331(6158). CrossRef
- Mitochondrial tRNA Mutations: Clinical and Functional Perturbations Zifa Emily, Giannouli Stamatina, Theotokis Paschalis, Stamatis Costas, Mamuris Zissis, Stathopoulos Constantinos. RNA Biology.2007;4(1). CrossRef
- Mitochondrial D310 instability in Asian Indian breast cancer patients Alhomidi Mohammed A., Vedicherla Bhavani, Movva Sireesha, Rao Prabhakar K., Ahuja Yog R., Hasan Qurratulain. Tumor Biology.2013;34(4). CrossRef
- Mitochondrial tRNA Import and Its Consequences for Mitochondrial Translation Schneider André. Annual Review of Biochemistry.2011;80(1). CrossRef
- Emerging roles of tRNA in adaptive translation, signalling dynamics and disease Kirchner Sebastian, Ignatova Zoya. Nature Reviews Genetics.2014;16(2). CrossRef
- Genetic, pathogenetic, and phenotypic implications of the mitochondrial A3243G tRNALeu(UUR) mutation Finsterer J.. Acta Neurologica Scandinavica.2007;116(1). CrossRef
- Homoplasmic MELAS A3243G mtDNA mutation in a colon cancer sample Lorenc Anna, Bryk Jarosław, Golik Paweł, Kupryjańczyk Jolanta, Ostrowski Jerzy, Pronicki Maciej, Semczuk Andrzej, Szołkowska Małgorzata, Bartnik Ewa. Mitochondrion.2003;3(2). CrossRef
- Renal cell carcinoma in a pediatric patient with an inherited mitochondrial mutation Sangkhathat Surasak, Kusafuka Takeshi, Yoneda Akihiro, Kuroda Seika, Tanaka Mio, Sakai Norio, Fukuzawa Masahiro. Pediatric Surgery International.2005;21(9). CrossRef
- Profile of breast cancer patients at a tertiary care hospital in north India Sandhu DS, Sandhu S, Karwasra RK, Marwah S. Indian Journal of Cancer.2010;47(1). CrossRef
- SEER cancer statistics review, 1975–2009 (vintage 2009 populations). Bethesda, MD: National Cancer Institute Howlader N, Noone A, Krapcho M, Neyman N, Aminou R, Altekruse S, et al . 2012.
- Cancer incidence and mortality: trends in the United Kingdom and constituent countries, 1993 to 2004 Westlake S, Cooper N. Health statistics quarterly / Office for National Statistics.2008;38:33-46.
- Report on trends of incidence (1970–2002) of and mortality (1952–2002) from cancer in Germany Becker Nikolaus, Altenburg Hans-Peter, Stegmaier Christa, Ziegler Hartwig. Journal of Cancer Research and Clinical Oncology.2006;133(1). CrossRef
- ABC of breast diseases: Screening for breast cancer Blamey R W. BMJ.2000;321(7262). CrossRef
- Clinical Value of Mitochondrial Mutations in Colorectal Cancer Lièvre Astrid, Chapusot Caroline, Bouvier Anne-Marie, Zinzindohoué Franck, Piard Françoise, Roignot Patrick, Arnould Laurent, Beaune Philippe, Faivre Jean, Laurent-Puig Pierre. Journal of Clinical Oncology.2005;23(15). CrossRef
- Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer Tseng Ling-Ming, Yin Pen-Hui, Chi Chin-Wen, Hsu Chih-Yi, Wu Chew-Wun, Lee Liang-Ming, Wei Yau-Huei, Lee Hsin-Chen. Genes, Chromosomes and Cancer.2006;45(7). CrossRef
- Analysis Association between Mitochondrial Genome Instability and Xenobiotic Metabolizing Genes in Human Breast Cancer Pavicic Walter H., Laguens Martin, Richard Silvina M.. Molecular Medicine.2009;15(5-6). CrossRef
- The Pathogenic A3243G Mutation in Human Mitochondrial tRNALeu(UUR)Decreases the Efficiency of Aminoacylation† Park Hyejeong, Davidson Edgar, King Michael P.. Biochemistry.2003;42(4). CrossRef
- Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid Zhu W.. Carcinogenesis.2004;26(1). CrossRef
- Somatic mutations of mitochondrial genome in hepatocellular carcinoma Yin Pen-Hui, Wu Cheng-Chung, Lin Jin-Ching, Chi Chin-Wen, Wei Yau-Huei, Lee Hsin-Chen. Mitochondrion.2010;10(2). CrossRef
- Learning from oncocytic tumors: Why choose inefficient mitochondria? Gasparre Giuseppe, Romeo Giovanni, Rugolo Michela, Porcelli Anna Maria. Biochimica et Biophysica Acta (BBA) - Bioenergetics.2011;1807(6). CrossRef
- Facile Detection of Mitochondrial DNA Mutations in Tumors and Bodily Fluids Fliss M. S.. Science.2000;287(5460). CrossRef
- Human mitochondrial tRNAs in health and disease Florentz C., Sohm B., Tryoen-T-th P., P-tz J., Sissler M.. Cellular and Molecular Life Sciences (CMLS).2003;60(7). CrossRef
- Mitochondrial tRNA mutations and disease Yarham John W., Elson Joanna L., Blakely Emma L., McFarland Robert, Taylor Robert W.. Wiley Interdisciplinary Reviews: RNA.2010;1(2). CrossRef
- A novel mitochondrial tRNA Arg mutation resulting in an anticodon swap in a patient with mitochondrial encephalomyopathy Roos Sara, Darin Niklas, Kollberg Gittan, Andersson Grönlund Marita, Tulinius Mar, Holme Elisabeth, Moslemi Ali-Reza, Oldfors Anders. European Journal of Human Genetics.2012;21(5). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details