C-Fos Digital Expression Analysis in Human PapillomavirusRelated Oral Squamous Cell Carcinoma
Download
Abstract
Background: Fos Proto-Oncogene (c-Fos) represents a well analyzed gene involved in solid malignancies’ development and progression. The corresponding protein forms heterodimer with c-jun, a strong transcription factor. C-Fos/c-Jun complex influences critically the intracellular signal transduction to the nucleus. Our aim was to detect and evaluate c-Fos protein expression patterns in oral squamous cell carcinomas (OSCC) tissues.
Materials and Methods: Fifty (n=50) formalin-fixed, paraffin-embedded primary OSCCs tissue sections were used. Immunohistochemistry and digital image analysis were implemented for identifying and evaluating c-Fos protein expression levels, respectively.
Results: C-Fos protein over expression (moderate to high imunostaining intensity values) was observed in 28/50 (56%) tissue cores, whereas low expression rates were detected in the rest of the examined cases (22/50- 44%). C-Fos overall expression was strongly associated with the stage and grade of the examined tumors (p=0.014, p=0.003, respectively) and also with Human papillomavirus (HPV) persistent infection (p=0.004). c-Fos up regulation is frequently observed in OSCCs.
Conclusion: C-Fos high expression levels are correlated with an aggressive phenotype (advanced stage/lymph node metastasis) in patients with OSCC, especially in HPV positive cases, especially High Risk subtypes. Due to its elevated oncogenic activity, c-Fos should be a target for novel therapeutic strategies in OSCC combined or not with other oncogenes involving in signaling transduction pathways.
Introduction
Solid malignancies demonstrate molecular profiles based on oncogenes’ over activation combined with suppressor genes silence. Concerning oncogenes, Fos Proto- Oncogene or AP-1 Transcription Factor Subunit (c-Fos) represents a critical gene involved in a variety of malignant tumours’ development and progression, including oral squamous cell carcinoma (OSCC). The Fos superfamily comprises c-Fos, FosB, FosL1, and FosL2 genes. c-Fos is a proto-oncogene that is the human homolog of the retroviral oncogene v-fos (gene locus: 14q24.3). It was initially analyzed and cloned in rat fibroblasts as the transforming gene of Finkel–Biskis–Jinkins murine osteogenic sarcoma virus [1]. The gene encodes a 62 kDa protein (380 amino acids), forming heterodimer with c-jun, a strong transcription factors), resulting in the formation of AP-1 (Activator Protein-1) complex. C-Fos/c-Jun complex influences intracellular signal transduction to the nucleus. c-Fos protein is implicated in critical cell functions including differentiation, proliferation, survival and also tissue homeostasis affected by hypoxia and angiogenesis [2]. Concerning OSCC, c-Fos aberrant expression seems to be a frequent event, especially in sub groups of patients associated on not with their corresponding clinico-histological features [3-4]. In the current study, we analyzed c- Fos protein expression levels in OSCC tissue sections by implementing a digital image analysis protocol on immunostined slides. To our knowledge there are very limited published data regarding the current methodology in OSCC oncoproteins’ analyses.
Materials and Methods
Study group
For the purposes of our study, fifty (n=50) archival, formalin-fixed and paraffin-embedded tissue specimens of histologically confirmed primary OSCC were used. Selection of the cases was based predominantly on the criterion of an aggressive phenotype as it is expressed due to advanced Grade and Stage histo-pathological features. The hospital ethics committee consented (Reference ID Research Protocol: 2226/09.09.2018) to the use of these tissues for research purposes, according to World Medical Association Declaration of Helsinki guidelines (2008, revised 2014). The tissue samples were fixed in 10% neutral-buffered formalin. Hematoxylin and eosin (H&E)-stained slides of the corresponding samples were reviewed for confirmation of histopathological diagnoses. All lesions were classified according to the histological typing criteria of World Health Organization (WHO) (5). Concerning HPV DNA status (positivity or not), the corresponding information was derived from patients’ medical file records. Among them, 18 HPV DNA positive cases were recorded. HPV 16/31/53 High Risk (HR) subtypes were detected mainly by analyzing the corresponding cases. Clinicopathological data of the examined cases are demonstrated in Table 1.
| Clinicopathological parameters | c-Fos | p value | |
| OSCC (n=50) | OE | LE | |
| 28/50 (56 %) | 22/50 (44 %) | ||
| n (%) | n | n | |
| Gender | 0.359 | ||
| Male | 44 (88) | 24/50 (48) | 20/50 (40) |
| Female | 6 (12) | 4/50 (8) | 2/50 (4) |
| HPV history | 0.004 | ||
| Positive | 18 (36) | 15/50 (30) | 3/50 (6) |
| Negative | 32 (64) | 13/50 (26) | 19/50 (38) |
| Grade | 0.003 | ||
| 1 | 18 (18) | 5/50 (10) | 13/50 (26) |
| 2 | 21 (58) | 13/50 (26) | 8/50 (16) |
| 3 | 11 (24) | 10/50 (20) | 1/50 (2) |
| Stage | 0.014 | ||
| I | 9 (18) | 3/50 (6) | 6/50 (12) |
| II | 26 (52) | 12/50 (24) | 14/50 (28) |
| III | 15 (30) | 13/50 (26) | 2/50 (4) |
| Smoking status | 0.208 | ||
| Current | 38 (76) | 22/50 (44) | 16/50 (32) |
| Former | 12 (24) | 6/50 (12) | 6/50 (12) |
OSCC, oral squamous cell carcinomas; OE, overexpression (moderate to high expression) staining intensity; values ≤142 on the examined stained nuclei; LE, low expression staining intensity values >156 on the examined stained nuclei
Antibodies and immunohistochemistry assay (IHC)
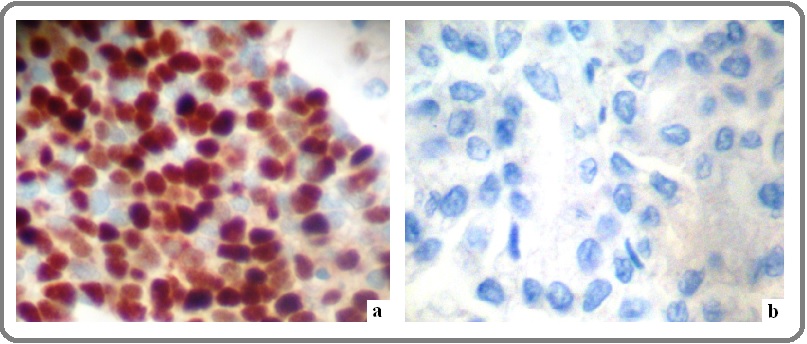
For the purposes of our study, we selected and applied the mouse monoclonal anti- c-Fos mouse monoclonal (clone CF2, Novocastra, Leica Biosystems, Newcastle, UK; dilution 1:40). IHC protocol for the antigen detection was carried out on a 4 μm thick paraffin sections of the current blocks. Tissue sections initially deparaffinized in xylene and rehydrated via graded ethanol - was immunostained according to the EN Vision + (DAKO, Denmark) assay using an automated staining system (I 6000 - Biogenex, CA, USA) and according to the corresponding antibodies manufacturer’s instructions. This specific assay is based on a soluble, dextran- polymer system preventing endogenous biotin reaction and increasing the quality of the stained slides. Briefly, the sections, after peroxidase blocking, were incubated with primary antibody for 30 min at room temperature and then incubated with Horseradish peroxidise labeled polymer-HRP LP for 30 min. A wash with TBS was performed. The antigen - antibody reaction was visualized using 3-3, diaminobenzidine tetrahydrocloride (DAB) as a chromogen substrate (8 min at room temperature). Finally, the tissue sections were slightly counterstained with hematoxylin for 30 sec, dehydrated and mounted. For negative control slides, the primary antibody was omitted. Nuclear predominantly but also peri-nuclear/cytoplasmic staining pattern was considered to be acceptable for the marker. Normal (non-cancerous) skin tissue sections demonstrating c-Fos expression was used as positive markers for its immunostaining pattern (Figure 1a,b).
Figure 1. C-Fos Expression Patterns on OSCC Tissue Sections. a. Over expression of c-Fos. Note nuclear mainly dense brown staining pattern (DAB stain, original magnification: 400x) b. Loss of expression (DAB stain, original magnification: 400x).
Digital Image Analysis assay (DIA)
C-fos protein expression levels were evaluated quantitatively by calculating the corresponding staining intensity levels (densitometry evaluation) in the immunostained malignant cells. We performed DIA using a semi-automated system (hardware: Microscope CX-31, Olympus, Melville, NY, USA, Digital camera, Sony, Tokyo, Jp; Windows XP/NIS-Elements Software AR v3.0, Nikon Corp, Tokyo, Japan). Areas of interest per tissue section were identified (5 optical fields at ×400 magnification) and filed in a digital database as snapshots. Measurements were performed by implementing a specific macro (mainly nuclear for tumor cells, according to manufacturer’s datasheet for monoclonal mouse anti-c- fos, Clone CF2, Novocastra, Leica Biosystems Newcastle Ltd, UK). Based on an algorithm, normal tissue sections (control) were measured independently and compared to the corresponding values in malignant tissue sections. A broad spectrum of continuous grey scale values (0-255) at the RedGreenBlue (RGB) analysis was available for detecting and discriminating different protein expression levels (Figure 2).
Figure 2. C-Fos Progressive Digital Image Analysis on OSCC Tissue Sections. a. Over expression of c-Fos. Note nuclear mainly and peri-nuclear/cytoplasmic brown staining pattern (DAB stain, original magnification: 100x) b. Red spots represent different expression values of c-Fos expression c. Green loops surrounding red spots represent the final stage of digital analysis providing numerical data (staining intensity values -DAB stain, original magnification: 100x).
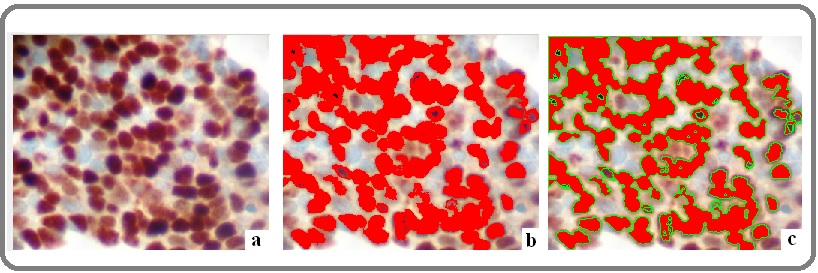
Immunostaining intensity values decreasing to 0 represent a progressive over expression of the marker, whereas values increasing to 255 show a progressive loss of its staining intensity. Total results and DIA values are demonstrated in Table 1.
Statistical analysis
Statistics software package IBM SPSS v25 (SPSS Inc, Chicago, IL, USA) was implemented. Associations between variables were assessed with of Pearson Chi- Square (χ2) test and Fisher’s exact. Correlation analysis with Spearman Rank test was performed for variables with significant chi2 associations. Two-tailed p-values ≤0.05 were considered statistically significant. Results and correlations (p-values) are described in Table 1.
Results
According to DIA implementation, all of the examined cases demonstrated c- Fos expression in different levels. C-Fos protein over expression (moderate to high imunostaining intensity values) was observed in 28/50 (56%) tissue sections, whereas low expression rates were detected in the rest of the examined cases (22/50- 44%). C-Fos protein over expression (moderate to high imunostaining intensity values) was observed in 28/50 (56%) tissue cores, whereas low expression rates were detected in the rest of the examined cases (22/50- 44%). C-Fos overall expression was strongly associated with the stage and grade of the examined tumors (p=0.014, p=0.003, respectively) and also with HPV persistent infection (p=0.004). No other statistical correlations were identified regarding the other clinic-pathological parameters (gender: p=0.359, smoking status: p=0.208).
Discussion
Oral cavity carcinoma represents a major proportion of Head and Neck Squamous Cell Carcinoma (HNSCC). OSCCs demonstrate an aggressive phenotype due to their increased capability to locally metastasize combined with distant lymph node metastases [6]. In solid malignancies deregulated transcriptional factors are correlated with aberrant gene expression [7]. Concerning OSCC, c-Fos over activation seems to be associated with other transcription factors (c-Jun/Fra-1) deregulation [8-9]. In fact, their high co-expression is correlated with poor prognosis in the corresponding patients. Besides these transcriptional factors, the role of c-Fos/mutant p53 protein in OSCC is under investigation. A study group analyzed their coexpression exploring also the involvement of another nucleolar protein (nucleophosmin) in their rise and progression [10]. They reported that high c-Fos/p53 expression levels lead to nucleophosmin up-regulation as a potential synergetic action of these molecules.
In the current study, we analyzed c-Fos protein expression by IHC on OSCC tissue sections measuring the levels of its staining intensity by implementing a DIA protocol. C-Fos up regulation (moderate to high nuclear/peri-nuclear cytoplasmic protein expression levels) was detected in a significant proportion of the examined cases. Overall protein expression was found to be correlated with the stage and also with HPV positivity. Concerning HPV-mediated carcinogenesis in oral mucosa, some molecular studies detected specific HPV-positive signatures that affect signal transduction pathways. In one of them, the role of HPV persistent infection in activation of AP-1, NF-κB, and STAT3 genes was explored. The study group concluded that there is a potential involvement of them in HPV-positive OSCC leading also to c-Fos aberrant expression [11]. Similarly, another experimental cell culture-based study focused on the influence of HPV16 E6 oncogene in c-Fos deregulation. The researchers reported that besides AP-1, c-Fos protein expression is increased by TGF-α in HPV16E6 positive cases [12]. Involvement of growth factors, such as TGF-a in HPV-dependent OSCC should lead to new therapeutic approaches in subsets of patients characterized by specific gene signatures. Similarly, induced apoptotic activity in OSCC is a major issue for research. According to apoptosis analyses in these malignancies, a study suggested that HOX-PBX complex inhibition by a specific agent (HXR9 peptide) is correlated also with c-Fos up regulation [13]. Furthermore, co-analysis of c-Fos/c- Jun, and also p53 immunohistochemistry led to a significant association with lymph node metastasis, poor differentiation and clinical stage of the examined OSCC tissues. The study group suggested this co-expression as a potential independent prognostic factor for overall survival in the corresponding patients [14].
In conjunction to the previously described results, another study detected over activation of c-Fos in invasive OSCC compared to adjacent non-malignant epithelia. A combination of nuclear and peri-nuclear cytoplasmic diffuse immunostaining was observed especially in cases demonstrated lymph node metastasis implicating also CD44-depended signal transduction pathway in patients with advanced stage [15]. Additionally, the role of another signal transduction pathway (Notch) in c-Fos over activation is under investigation. A study group showed that targeting overexpressed Notch genes - including JAG1/ JAG2- in OSCC by applying γ-secretase molecule, c-Fos m RNA levels were also decreased [16]. Novel molecular technologies in OSCC gene screening based on c-DNAs microarrays have also detected distinct patterns of gene expression in different sub-group of patients, including oncogenes [17]. Concerning c-fos protein expression evaluation, we implemented a DIA protocol that provides an objective, accurate and fast mean for measuring the immunostaining intensity levels. According to our published experience in a variety of biomarkers, DIA is a useful tool for research and diagnostic reasons -under specific terms- enhancing the innovative evidence- based medicine [18-22].
In summary, c-Fos is a critical gene frequently up regulated in OSCCs. C-Fos high expression levels are correlated with an aggressive phenotype (advanced stage/ lymph node metastasis) in patients with OSCC. HR HPV- mediated carcinogenesis in a subset of cases is associated also with increased c-Fos protein expression. Due to its elevated oncogenic activity, c-Fos should be a target for novel therapeutic strategies in OSCC combined or not with other oncogenes involving in signalling transduction pathways.
Acknowledgments
Authors acknowledge the significant scientific contribution of George Vilaras, technologist in the Department of Pathology, Medical School, University of Athens, Greece, as an expert in IHC/ICC/CISH techniques.
Sources of funding
None.
Declaration of competing interest
None.
References
- FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. Curran T, Peters G, Van Beveren C, Teich N M, Verma I M. Journal of Virology.1982;44(2). CrossRef
- Assessment of c-Jun, c-Fos and cyclin D1 in premalignant and malignant oral lesions Turatti Eveline, da Costa Neves Adriana, de Magalhães Marina Helena Cury Gallottini, de Sousa Suzana Orsini Machado. Journal of Oral Science.2005;47(2). CrossRef
- Differential Expression of c-fos Proto-Oncogene in Normal Oral Mucosa versus Squamous Cell Carcinoma Krishna Akhilesh, Bhatt Madan Lal Brahma, Singh Vineeta, Singh Shraddha, Gangwar Pravin Kumar, Singh Uma Shankar, Kumar Vijay, Mehrotra Divya. Asian Pacific Journal of Cancer Prevention.2018;19(3). CrossRef
- Transactivation and expression patterns of Jun and Fos/AP-1 super-family proteins in human oral cancer Mishra Alok, Bharti Alok C., Saluja Daman, Das Bhudev C.. International Journal of Cancer.2009. CrossRef
- World health organization classification of head and neck tumours El-Naggar AK, Chan JKC, Grandis JR, et al . 4th ed. Lyon: IARC Press.2017.
- Genetic etiology of oral cancer Ali Johar, Sabiha Bibi, Jan Hanif Ullah, Haider Syed Adnan, Khan Abid Ali, Ali Saima S.. Oral Oncology.2017;70. CrossRef
- Hallmarks of Cancer: The Next Generation Hanahan Douglas, Weinberg Robert A.. Cell.2011;144(5). CrossRef
- Prognostic value from integrative analysis of transcription factors c-Jun and Fra-1 in oral squamous cell carcinoma: a multicenter cohort study Xu Hao, Jin Xin, Yuan Yao, Deng Peng, Jiang Lu, Zeng Xin, Li Xiao-Song, Wang Zhi-Yong, Chen Qian-Ming. Scientific Reports.2017;7(1). CrossRef
- Selective participation of c-Jun with Fra-2/c-Fos promotes aggressive tumor phenotypes and poor prognosis in tongue cancer Gupta Shilpi, Kumar Prabhat, Kaur Harsimrut, Sharma Nishi, Saluja Daman, Bharti Alok C., Das Bhudev C.. Scientific Reports.2015;5(1). CrossRef
- Oncogene c‐fos and mutant R175H p53 regulate expression of Nucleophosmin implicating cancer manifestation Senapati Parijat, Dey Suchismita, Sudarshan Deepthi, Das Sadhan, Kumar Manoj, Kaypee Stephanie, Mohiyuddin Azeem, Kodaganur Gopinath S., Kundu Tapas K.. The FEBS Journal.2018;285(18). CrossRef
- Characterization of key transcription factors as molecular signatures of HPV-positive and HPV-negative oral cancers Verma Gaurav, Vishnoi Kanchan, Tyagi Abhishek, Jadli Mohit, Singh Tejveer, Goel Ankit, Sharma Ankita, Agarwal Kiran, Prasad Subhash Chandra, Pandey Durgatosh, Sharma Shashi, Mehrotra Ravi, Singh Sukh Mahendra, Bharti Alok Chandra. Cancer Medicine.2017;6(3). CrossRef
- HPV16E6-Dependent c-Fos Expression Contributes to AP-1 Complex Formation in SiHa Cells Liang Feixin, Kina Shinichiro, Takemoto Hiroyuki, Matayoshi Akira, Phonaphonh Thongsavanh, Sunagawa Nao, Arakaki Keiichi, Arasaki Akira, Kuang Hai, Sunakawa Hajime. Mediators of Inflammation.2011;2011. CrossRef
- Targeting HOX-PBX interactions causes death in oral potentially malignant and squamous carcinoma cells but not normal oral keratinocytes Platais Christopher, Radhakrishnan Raghu, Niklander Ebensperger Sven, Morgan Richard, Lambert Daniel W., Hunter Keith D.. BMC Cancer.2018;18(1). CrossRef
- Combined Expression of c-jun, c-fos, and p53 Improves Estimation of Prognosis in Oral Squamous Cell Carcinoma Wang Shan, Xu Xin, Xu Fei, Meng Yan, Sun Changsheng, Shi Lei, Zhao Eryang. Cancer Investigation.2016;34(8). CrossRef
- Overexpression of c-fos promotes cell invasion and migration via CD44 pathway in oral squamous cell carcinoma Dong Cong, Ye Dong-Xia, Zhang Wen-Bin, Pan Hong-Ya, Zhang Zhi-Yuan, Zhang Lei. Journal of Oral Pathology & Medicine.2014;44(5). CrossRef
- Expression and influence of Notch signaling in oral squamous cell carcinoma Osathanon Thanaphum, Nowwarote Nunthawan, Pavasant Prasit. Journal of Oral Science.2016;58(2). CrossRef
- Gene expression analysis by cDNA microarray in oral cancers from two Western populations Lunde ML, Warnakulasuriya S, Sand L, et al . Anticancer Res.2010;30:1083-1091.
- Digital Analysis of BCL2 Expression in Laryngeal Squamous Cell Carcinoma CHRYSOVERGIS ARISTEIDIS, PAPANIKOLAOU VASILEIOS S., TSIAMBAS EVANGELOS, RAGOS VASILEIOS, PESCHOS DIMITRIOS, KYRODIMOS EFTHYMIOS. Anticancer Research.2019;39(3). CrossRef
- Topoisomerase I deregulation in laryngeal squamous cell carcinomas based on tissue microarray analysis Papadas TA, Tsiambas E, Mastronikolis NS, et al . JBUON.2017;22:771-776.
- Combined EGFR/ALK Expression Analysis in Laryngeal Squamous Cell Carcinoma POLITI ANASTASIA, TSIAMBAS EVANGELOS, MASTRONIKOLIS NICHOLAS S., PESCHOS DIMITRIOS, ASPROUDIS IOANNIS, KYRODIMOS EFTHYMIOS, ARMATA ILIANNA E., CHRYSOVERGIS ARISTEIDIS, ASIMAKOPOULOS ASIMAKIS, PAPANIKOLAOU VASILEIOS S., BATISTATOU ANNA, RAGOS VASILEIOS. In Vivo.2019;33(3). CrossRef
- Targeting topoisomerase IIa in endometrial adenocarcinoma: a combined chromogenic in situ hybridization and immunohistochemistry study based on tissue microarrays TSIAMBAS E., ALEXOPOULOU D., LAMBROPOULOU S., GERONTOPOULOS K., KARAKITSOS P., KARAMERIS A.. International Journal of Gynecological Cancer.2006;16(3). CrossRef
- Chromogenic In Situ Hybridization Analysis of Epidermal Growth Factor Receptor Gene/Chromosome 7 Numerical Aberrations in Hepatocellular Carcinoma Based on Tissue Microarrays Tsiambas Evangelos, Manaios Loukas, Papanikolopoulos Costas, Rigopoulos Dimitrios N., Tsounis Dimitrios, Karameris Andreas, Soultati Aspasia, Koliopoulou Antigoni, Kravvaritis Christos, Sergentanis Theodoros, Patsouris Efstratios, Dourakis Spyridon. Pathology & Oncology Research.2009;15(3). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details