Prevalence of BRAF V600E Mutation in the Iranian Patients with Hairy Cell Leukemia: A Retrospective Study
Download
Abstract
Objective: BRAF V600E mutation has several implications in hairy cell leukemia (HCL). The prevalence of This mutation has been investigated in various populations, but not in Iran. In this study, we evaluated the prevalence of BRAF V600E mutation in an Iranian HCL population as well as its association with the patients’ characteristics.
Methods: In a retrospective (archival) study, 20 HCL patients with the confirmed immunophenotypic and morphologic diagnosis were included. Paraffin-embedded blocks of bone marrow aspirate were used to investigated BRAF V600E mutation using amplification refractory mutation system (ARMS) PCR. Demographic, clinical, laboratory, and immunophenotypic characteristics of patients were extracted from the patients medical profiles.
Result: BRAF V600E mutation was present in 17 (85%) HCL patients and absent in three (15%) patients. The mean age of the patients was 44.76 ± 8.69 years in mutation-positive and 62.33 ± 8.69 in mutation-negative patients. This difference was statistically significant (p=0.013). No significant difference was found between the laboratory indices of the mutation-positive and mutation-negative groups. The clinical, morphologic, and immunophenotypic characteristics of the two groups were also statistically comparable.
Conclusion: BRAF V600E mutation is present in the majority of the Iranian HCL patients and is associated with younger age of presentation.
Introduction
Hairy cell leukemia (HCL) is a rare and slow-growing hematological malignancy with unknown etiology, accounting for 2% of all leukemias. It is a very heterogeneous group of B‐cell disorders characterized by excessive B-cell proliferation in the bone marrow with a hairy look under the microscope. The patients are generally presented with pancytopenia and splenomegaly, as well as non-specific clinical symptoms such as fatigue and weakness [1]. Although the etiology of HCL is not well identified, a connection between specific gene mutations has been elucidated [2].
MAP kinase cascade is a key signaling pathway that mediates a wide variety of cellular functions, including cellular growth, proliferation, differentiation, and apoptosis. Activation of the MAP kinase pathway is a frequent event in tumorigenesis. The BRAF gene is a human gene with different variant products involved in the MAPK pathway and its mutation might lead to uncontrolled cell proliferation. About 43 mutations have been identified in the BRAF gene, which has been attributed to a variety of human malignancy [3-4].
V600E is the most common mutation of BRAF (BRAF V600E), which occurs on exon 15 and results in the alteration of amino acid 600 from valine to glutamate in the BRAF protein, followed by continuous activation of the downstream kinases and increased cell proliferation [5]. BRAF V600E has been detected in various cancers with different prevalence including colorectal cancers (about 10% of cases), non–small cell lung cancers (about 7% of cases), papillary thyroid cancers (about 51% of cases), and melanoma (about 50% of cases) [6].
Although BRAF V600E mutation has been identified in over 97% of the HCL cases so far investigated [7], a lower frequency of BRAF mutations (79%) has also been reported [8]. Considering the implications of BRAF V600E mutation for the pathogenesis, diagnosis, prognosis, and targeted therapy of HCL, evaluation of its prevalence in different HCL populations is of critical importance [4]. However, the prevalence of BRAF V600E mutation in the Iranian HCL population has not been studied in earlier investigations. In this multi-center study, we evaluate the prevalence of BRAF V600E mutation in Iranian HCL patients.
Materials and Methods
Study design and data collection
This retrospective (archival) study was approved by the review board of our institute. The target population was consecutive HCL patients who were referred to the oncology hospitals of Shiraz, Iran, between 2012 and 2015. Patients with a confirmed immunophenotypic and morphologic diagnosis of HCL were included, providing that their paraffin-embedded block of bone marrow aspirate was available for genetic evaluation. Finally, 20 patients were identified as eligible for the study. Demographic, clinical, laboratory, and immunophenotypic characteristics of patients were extracted from the patients medical profiles.
Evaluation of BRAF V600E mutation
After deparaffinization of paraffin-embedded sections, DNA was extracted using GeNet Bio kit (GeNet Bio inc. Daejeon, South Korea). The quality and quantity of DNA were assessed with a nanodrop-1000 spectrophotometer. Then DNA amplification was done using amplification refractory mutation system-polymerase chain (ARMS- PCR) as earlier described [9]. The ARMS-PCR primer sequences are demonstrated in Table 1.
| Forward (F) | 5’ –CTC TTC ATA ATG CTT GCT CTG ATA G-3’ |
| Reverse (R) | 5’-GCCTCAATTCTTACCATCCAC-3’ |
| Forward wild-type identifying (Fwi) | 5’-GTGATTTTGGTCT AGCTACAGT-3’ |
| Reverse mutation identifying: (Rmi) | 5’-CCCACTCCATCGAGATTTCT-3’ |
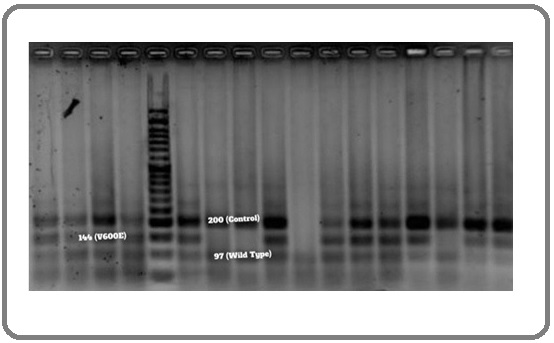
Four PCR primers were included in one PCR tube. Accordingly, F-R primer pair amplifies a common 200 bp fragment flanking the site of mutation. F-Rmi primer pair amplifies a fragment of 144 bp specific to the BRAF V600E mutation. Fwi-R primer pair amplifies a 97 bp fragment specific to wild-type BRAF gene.
PCR was performed in a 25 μl final volume, which included 1 × Buffer, 30 ng extracted DNA template, 2 mM MgCl2, 1 unit of Hotstar Taq DNA polymerase (Qiagen Science, Valencia, CA), 200 μM of dNTPs, 400 nM primer F, 200 nM primer R, 200 nM primed Fwi, and 800 nM primer Rmi. PCR was started with denaturation at 95°C for 5 min, followed by 40 cycles of 94°C for 20 sec, 62°C for 20 sec and 72°C for 20 sec, and a final extension step at 72°C for 5 min. PCR products were analyzed by 2.5% agarose gel electrophoresis and were visualized under UV light (Figure 1).
Figure 1. Evaluation of PCR Products on the Agarose Gel Electrophoresis under UV Light.
Statistical analysis
SPSS for Windows version 16 (SPSS Inc., Chicago, Ill., USA) was used for the statistical analysis of the data. Descriptive statistics were demonstrated with mean ± standard deviation or number & percentage. A Mann–Whitney U test was used to compare quantitative variables across two groups. A chi-square test was used to evaluate the statistical association between categorical variables. A p-value of fewer than 0.05 was considered statistically significant.
Results
The study population included 19 (95%) males and one (5%) female with a mean age of 47.4±10.3 years (range 28-66). BRAF V600E mutation was positive in 17 (85%) patients and negative in three (15%) patients. The mean age of the patients was significantly lower in patients with positive BRAF V600E mutation (p=0.013). No other significant association was found between the patients with and without BRAF V600E mutation. Laboratory indices were also statistically comparable between the two groups (Table 2).
| Variables | BRAF Gene Mutation | ||
| Negative (n=3) | Positive (n=17) | P-value | |
| Sex | |||
| Male | 3 (100) | 16 (94.1) | 0.666 |
| Female | 0 | 1 (5.9) | |
| Age (years) | 62.33 ± 8.69 | 44.76 ± 8.69 | 0.013* |
| WBC count (/µL) | 2733 ± 2020 | 2241 ± 868 | 0.468 |
| Neutrophil (%) | 50 ± 18 | 39 ± 18 | 0.356 |
| Lymphocyte (%) | 50 ± 18 | 60 ± 18 | 0.364 |
| Hemoglobin (g/dL) | 9.8 ± 4.2 | 9.9 ± 1.7 | 0.91 |
| Platelet count (/µL) | 81000 ± 32908 | 55411 ± 26260 | 0.149 |
Data are presented as mean ± SD or number (%). P <0.05 is considered significant.
The mean size of the spleen was 18.7±2.3 cm in the negative and 17.4±3.5 cm in the positive BRAF V600E mutation group (p=0.416). Accordingly, splenomegaly was noticed in one patient of the positive BRAF V600E group and no patient of the negative BRAF V600E group (p=0.666). No other significance was also found between the clinical features of the two groups. Bone marrow morphology was statistically comparable between the two groups, as well (Table 3).
| Variables | BRAF gene mutation | ||
| Negative (n=3) | Positive (n=17) | P-value | |
| Hospital Admissions | |||
| · Once | 0 | 2 (11.8) | |
| ·Twice & More | 3 (100) | 12 (70.6) | 0.555 |
| · Never | 0 | 3 (17.6) | |
| Spleen size (cm) | 18.7±2.3 | 17.4±3.5 | 0.416 |
| Splenomegaly | |||
| · Yes | 0 | 1 (5.9) | 0.666 |
| · No | 3 (100) | 16 (94.1) | |
| Bone marrow Morphology | |||
| · Hypocellular | 2 (66.7) | 5 (29.4) | |
| ·Hypercellular | 0 | 3 (17.6) | 0.42 |
| · Mosaic-pattern | 1 (33.3) | 9 (53) |
Data are presented as mean ± SD or number (%). P <0.05 is considered significant.
Moreover, no significant difference was found between the immunophenotypic characteristics of the two groups (Table 4).
| Variables | BRAF gene mutation | P-value | |
| Negative (n=3) | Positive (n=17) | ||
| CD3 | |||
| · Positive | 1 (33.3) | 2 (18.2) | 0.571 |
| · Negative | 2 (66.7) | 9 (81.8) | |
| CD5 | |||
| · Positive | 0 | 2 (16.7) | 0.666 |
| · Negative | 2 (100) | 12 (83.3) | |
| CD7 | |||
| · Positive | 0 | 2 (100) | 0.083 |
| · Negative | 1 (100) | 0 | |
| CD8 | |||
| · Positive | 1 (100) | 2 (66.7) | 0.505 |
| · Negative | 0 | 1 (33.3) | |
| CD10 | |||
| · Positive | 0 | 2 (20) | 0.621 |
| · Negative | 1 (100) | 8 (80) | |
| CD11c | |||
| · Positive | 2 (100) | 16 (94.1) | 0.725 |
| · Positive focally | 0 | 1 (5.9) | |
| CD19 | |||
| · Positive | 2 (100) | 7 (87.5) | 0.598 |
| · Negative | 0 | 1 (12.5) | |
| CD20 | |||
| · Positive | 1 (33.3) | 11 (64.7) | 0.306 |
| · Positive diffusely | 2 (66.7) | 6 (35.3) | |
| FMC7 | |||
| · Positive | 0 | 1 (100) | 0.157 |
| · Negative | 1 (100) | 0 |
Data are presented as number (%). P <0.05 is considered significant.
Discussion
In this study, we evaluated the presence of BRAF V600E mutation in Iranian HCL patients in a metacentric study. We also investigated the association of this mutation with the patients’ characteristics. According to this study, BRAF V600E mutation was present in 85% of HCL patients. Patients with negative BRAF V600E mutation had significantly higher age. No significant association was found between the BRAF V600E mutation and characteristics of HCL patients such as laboratory indices, immunophenotypic markers, etc.
In earlier studies, various rates have been reported for BRAF V600E mutation in HCL patients. Tiacci et al. evaluated the BRAF V600E mutation in 47 Italian HCL patients. BRAF V600E mutation was present in all patients [4]. Boyd et al. reported 100% BRAF V600E mutation (n=48) in English HCL patients [10]. Arciani et al. assessed BRAF V600E mutation in 62 HCL patients. BRAF V600E mutation was detected in all cases [11]. Blombery et al. investigated BRAF V600E mutation in 51 Australian HCL patients (59 samples). In total, BRAF V600E mutation was detected in 36 out of 51 patients (70.5%) [12]. Bibi et al. detected BRAF V600E mutation in 89.1% (41/46 patients) of the Indian HCL population [11]. In the present study, we detected BRAF V600E mutation in 85% of Iranian HCL patients that was similar to that reported in the study of Bibi et al [11]. Nonetheless, the difference between studies could be attributed to the method of detection. Blombery et al. used high resolution melting analysis and confirmatory Sanger sequencing for the detection of BRAF V600E mutation. The detected mutations were not equal using the two methods, so that high resolution melting analysis detected mutation in 42 samples, which was confirmed by sequencing in 38 [12]. Therefore, the method of evaluation could be regarded as a source of heterogeneity between the studies.
Langabeer et al. evaluated the correlation of the BRAF V600E mutation with immunophenotypic characteristics of the 24 patients with a classic HCL. The CD11c+/CD20+/ CD25+/CD103+/FMC7+ HCL immunophenotype was detected in 23 HCL patients. BRAF V600E mutation was also detected in 23 patients. They suggested a high degree of correlation between the presence of BRAF V600E mutation and established diagnostic criteria and highlighted the value of a multifaceted approach to the diagnosis of HCL [13]. We could not detect the association between BRAF V600E mutation and immunophenotypic characteristics due to the high missing data in this section. In the study of Bibi et al. HCL patients with BRAF V600E mutation presented at a younger age. However, no significant difference was found between the other characteristic features of the patients with and without BRAF mutation, such as in laboratory parameters [14]. We also detected an association between BRAF V600E mutation and the age of the presentation, so that patients with BRAF V600E mutation were significantly younger. Similar to the study of Bibi et al. laboratory indices were not associated with BRAF V600E mutation in the present study.
The present study has several weaknesses. First, the PCR results were not confirmed by sequencing. Second, the number of patients in the negative BRAF mutation group was considerably small. Therefore, the power of statistical analysis could be poor. Finally, a significant number of data were missing, particularly in immunophenotypic characteristics. Therefore, future studies with larger patients’ numbers are required to confirm the results of this study.
In conclusion, BRAF V600E mutation was present in 85% of Iranian HCL patients and absent in 15%. The presence of the mutation was associated with the younger age of the patients, but not with other patients’ characteristics such as immunophenotypic and laboratory indices. Our finding suggests a diagnostic role for BRAF V600E mutation in the Iranian HCL population.
Acknowledgements
We would like to thank the Shiraz University of Medical Sciences for providing the infrastructure and implications for this study, and we would especially like to thank Mrs. Valibegi, a medical school technician, for all her efforts and support during this study.
Funding Statement
This thesis is supported by deputy dean of School of Medicine based on research project number 7008 dated 1392/1/1 and sponsored by deputy chancellor of Shiraz University of Medical Sciences.
Statement conflict of Interest
No potential conflict of interest was reported by the authors.
References
- Hairy cell leukemia 2018: Update on diagnosis, risk-stratification, and treatment Troussard Xavier, Cornet Edouard. American Journal of Hematology.2017;92(12). CrossRef
- Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations Durham Benjamin H., Getta Bartlomiej, Dietrich Sascha, Taylor Justin, Won Helen, Bogenberger James M., Scott Sasinya, Kim Eunhee, Chung Young Rock, Chung Stephen S., Hüllein Jennifer, Walther Tatjana, Wang Lu, Lu Sydney X., Oakes Christopher C., Tibes Raoul, Haferlach Torsten, Taylor Barry S., Tallman Martin S., Berger Michael F., Park Jae H., Zenz Thorsten, Abdel-Wahab Omar. Blood.2017;130(14). CrossRef
- BRAF mutation in hairy cell leukemia Ahmadzadeh Ahmad, Shahrabi Saeid, Jaseb Kaveh, Norozi Fatemeh, Shahjahani Mohammad, Vosoughi Tina, Hajizamani Saeideh, Saki Najmaldin. Oncology Reviews.2014. CrossRef
- BRAFMutations in Hairy-Cell Leukemia Tiacci Enrico, Trifonov Vladimir, Schiavoni Gianluca, Holmes Antony, Kern Wolfgang, Martelli Maria Paola, Pucciarini Alessandra, Bigerna Barbara, Pacini Roberta, Wells Victoria A., Sportoletti Paolo, Pettirossi Valentina, Mannucci Roberta, Elliott Oliver, Liso Arcangelo, Ambrosetti Achille, Pulsoni Alessandro, Forconi Francesco, Trentin Livio, Semenzato Gianpietro, Inghirami Giorgio, Capponi Monia, Di Raimondo Francesco, Patti Caterina, Arcaini Luca, Musto Pellegrino, Pileri Stefano, Haferlach Claudia, Schnittger Susanne, Pizzolo Giovanni, Foà Robin, Farinelli Laurent, Haferlach Torsten, Pasqualucci Laura, Rabadan Raul, Falini Brunangelo. New England Journal of Medicine.2011;364(24). CrossRef
- New insight into BRAF mutations in cancer Dhomen Nathalie, Marais Richard. Current Opinion in Genetics & Development.2007;17(1). CrossRef
- Improvements in Clinical Outcomes for BRAFV600E-Mutant Metastatic Colorectal Cancer Morris Van K., Bekaii-Saab Tanios. Clinical Cancer Research.2020;26(17). CrossRef
- BRAF V600E mutation in hairy cell leukemia: from bench to bedside Falini Brunangelo, Martelli Maria Paola, Tiacci Enrico. Blood.2016;128(15). CrossRef
- Both variant and IGHV4-34–expressing hairy cell leukemia lack the BRAF V600E mutation Xi Liqiang, Arons Evgeny, Navarro Winnifred, Calvo Katherine R., Stetler-Stevenson Maryalice, Raffeld Mark, Kreitman Robert J.. Blood.2012;119(14). CrossRef
- Sensitive detection of BRAF V600E mutation by Amplification Refractory Mutation System (ARMS)-PCR Huang Tiangui, Zhuge Jian, Zhang Wenyong W. Biomarker Research.2013;1(1). CrossRef
- High resolution melting analysis for detection of BRAF exon 15 mutations in hairy cell leukaemia and other lymphoid malignancies Boyd Elaine M., Bench Anthony J., van ‘t Veer Mars B., Wright Penny, Bloxham David M., Follows George A., Scott Mike A.. British Journal of Haematology.2011;155(5). CrossRef
- The BRAF V600E mutation in hairy cell leukemia and other mature B-cell neoplasms Arcaini Luca, Zibellini Silvia, Boveri Emanuela, Riboni Roberta, Rattotti Sara, Varettoni Marzia, Guerrera Maria Luisa, Lucioni Marco, Tenore Annamaria, Merli Michele, Rizzi Silvia, Morello Lucia, Cavalloni Chiara, Da Vià Matteo C., Paulli Marco, Cazzola Mario. Blood.2012;119(1). CrossRef
- Detection of BRAF mutations in patients with hairy cell leukemia and related lymphoproliferative disorders Blombery P. A., Wong S. Q., Hewitt C. A., Dobrovic A., Maxwell E. L., Juneja S., Grigoriadis G., Westerman D. A.. Haematologica.2011;97(5). CrossRef
- Correlation of the BRAF V600E mutation in hairy cell leukaemia with morphology, cytochemistry and immunophenotype LANGABEER S. E., O’BRIEN D., LIPTROT S., FLYNN C. M., HAYDEN P. J., CONNEALLY E., BROWNE P. V., VANDENBERGHE E.. International Journal of Laboratory Hematology.2012;34(4). CrossRef
- BRAFV600E mutation in hairy cell leukemia: A single-center experience Patkar Nikhil, Bibi Asma, Java Shrutika, Chaudhary Shruti, Joshi Swapnali, Mascerhenas Russel, Rabade Nikhil, Tembhare Prashant, Subramanian PapagudiGanesan, Gujral Sumeet, Menon Hari, Khattry Navin, Sengar Manju, Bagal Bhausaheb, Jain Hasmukh. Indian Journal of Pathology and Microbiology.2018;61(4). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details