Comprehensive Analysis of the Expression and Prognosis for DCTPP1 gene in Breast Cancer
Download
Abstract
Background: Breast cancer is a common malignancy in women. DCTPP1 is a potential target for the development of antitumor drugs, and plays an important role in the process of DNA replication. Aims: To investigate the biological role of DCTPP1 gene, as well as its expression in breast cancer and its relation to patient prognosis.
Materials and Methods: Breast cancer data was derived from the TCGA database. Using the UALCAN database, the expression level of DCTPP1 mRNA in breast cancer tissues was investigated. The expression of DCTPP1 in various pathological types of breast cancer was studied using the Human Protein Atlas. UALCAN was also used to investigate the relationship between DCTPP1 gene expression and breast cancer patient prognosis. Bioinformatics studied the proteins related to DCTPP1 expression and their roles in the GeneMANIA and WebGestalt databases.
Results: DCTPP1 mRNA was significantly expressed in breast cancer compared to normal breast tissue (P<0.001). DCTPP1 was shown to be highly expressed in breast cancer tissues from different pathological types and stages (P<0.001). The DCTPP1 protein was expressed at a higher frequency in breast cancer than in normal breast tissue. When compared to patients with low DCTPP1 expression, patients with high DCTPP1 expression had a considerably shorter overall survival time. The 20 proteins related to DCTPP1 expression were mostly located in the nucleus and membrane, and were involved in biological regulation, stimulus response, metabolic process, and other processes, according to gene ontology analysis. It plays an important role in protein binding, ion binding, and nucleic acid binding.
Conclusion: DCTPP1 is highly expressed in breast cancer, and is associated to a poor prognosis for patients with breast cancer. DCTPP1 may be a potential therapy and intervention target for breast cancer.
Introduction
Breast cancer is a common malignancy in women. There are about 2 million new breast cancer patients in the world every year, and the incidence of breast cancer is increasing year by year [1], which is a serious threat to women’s health. The United States and Northern Europe have a high incidence of breast cancer, while Asia has the lowest incidence rate worldwide [2].
Nucleoside triphosphate pyrophosphatase (NTP- PPase) can hydrolyze the phosphodiester bond of nucleoside triphosphate dNTP to form nucleoside monophosphate dNMP, and releasing pyrophosphate. The hydrolysis of abnormal nucleotides by NTP-PPase significantly reduces the abnormal nucleotides in the cell, avoids the incorporation of abnormal nucleotides in the DNA synthesis process, and improves the accuracy of DNA replication [3]. Deoxycytidine triphosphate pyrophosphatase 1 (DCTPP1) gene is a member of the NTP-PPase family with NTP-PPase activity. It has been reported that DCTPP1 is a potential target for the development of antitumor drugs [4,5], and it plays an important role in the process of DNA replication [6]. However, there are few reports on the expression and function of DCTPP1 in clinical breast cancer tissues. This study aims to use tumor-related databases to clarify the expression and distribution of DCTPP1 in breast cancer and its relationship with the prognosis of breast cancer patients, and to explore the function of DCTPP1 in the occurrence and development of breast cancer, to lay the foundation for the study of DCTPP1 gene in breast cancer.
Materials and Methods
The Cancer Genome Atlas
The data source of breast cancer patients was come from The Cancer Genome Atlas (TCGA, http://cancergenome. nih.gov/). The database includes basic clinical information of tumor cases, such as basic information, treatment progress, clinical stage, tumor pathology and survival status, including mRNA, microRNA, Mutation, Protein, Methylation information, etc. 1097 breast cancer tissues and 114 normal breast tissues were extracted using the databases mentioned below for further analysis. The gene expression data were collected from the fresh tissue samples.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a web-based collaborative resource based on level 3 RNA-seq and clinical evidence from the TCGA database for 31 cancer types. It can be used to compare the transcriptional expression of possible genes of interest in tumor and normal samples, as well as the relationship between transcriptional expression and clinicopathologic parameters [7].
The database was used to compare the expression level of the DCTPP1 mRNA between tumor tissues and normal tissue, and in different stages and pathological subtypes of breast cancer. We further investigated and analyzed the correlation between DCTPP1 gene expression level and the prognosis of breast cancer patients, compared the overall survival between patients with high versus low DCTPP1 expression.
The Human Protein Atlas
The Human Protein Atlas (https://www.proteinatlas. org/) is a database dedicated to storing human protein interaction information, the protein expression status of genes in tumor tissues and normal tissues could be queried by this database [8]. The DCTPP1 protein expression level in normal tissue, breast duct cancer tissue, and breast lobular cancer tissue were extracted and compared.
GeneMANIA
The GeneMANIA database (https://genemania.org/) is a flexible, user-friendly web interface, can provide protein-protein, protein-DNA and genetic interactions, pathways, physiological and biochemical reactions and other information [9]. The function of protein-protein interaction (PPI) is the main priority of cellular biology research and serves as a prerequisite for system biology. Proteins interact with other proteins within the cell to fulfill their functions, and information produced by a PPI network increases awareness of the protein’s role. Using this database, we constructed the PPI network, and determined key genes that interacted with the DCTPP1.
WebGestalt
WebGestalt (http://www.webgestalt.org/) is a web- based integrated data mining system, composed of four modules: gene set management, information retrieval, organization/visualization, and statistics [10]. The gene ontology (GO) term is used for gene product annotation, and it is divided into three categories: biological process, molecular function, and cellular component. The knowledge of molecular activity, cellular role, and the position in a cell where the genes perform their functions is the principal cause for defining GO words.
The key genes that interacted with the DCTPP1 gene would be used to perform GO analysis with WebGestalt to understand the cellular component, biological process, and molecular function that it participated in.
Statistical method
Kaplan-Meier survival analysis was used to calculate survival rate, log-rank test was used to estimate the difference in survival rate. A two-tailed P<0.05 was considered statistically significant.
Results
Differential expression of DCTPP1 in breast cancer and normal tissues
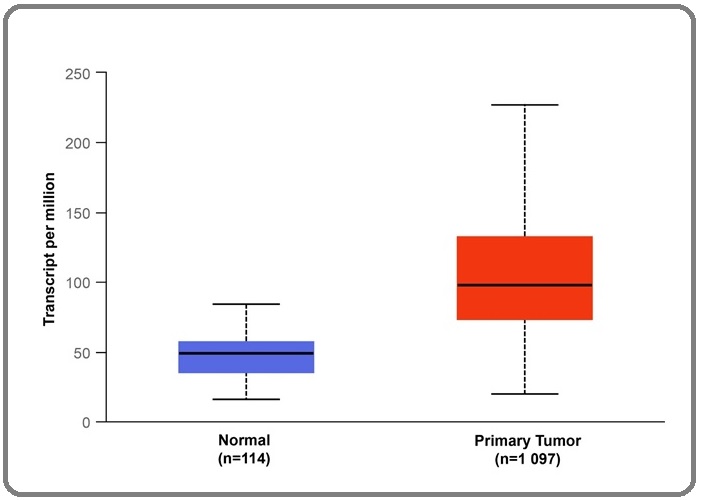
We compared the expression levels of DCTPP1 mRNA in 1097 breast lobular carcinoma tissues and 114 normal breast tissues using data from the UALCAN database. The median expression level of DCTPP1 mRNA in normal breast tissues was 48.7 (35.4-57.1) transcript per million (TPM), whereas in breast cancer tissues it was 97.5 (73.4-132.0) TPM, which was substantially higher than the expression level in normal breast tissue (P<0.001) (Figure 1).
Figure 1. The Expression Level of DCTPP1 mRNA in Breast Lobular Carcinoma Tissues and Normal Breast Tissues Based on the TCGA Database.
The expression level of DCTPP1 in different stages and pathological subtypes of breast cancer
Figure 2A indicated that, according to the clinical TNM stage, the DCTPP1 expression level in the tissues of breast lobular carcinoma patients with stage I, II, III, and IV was significantly higher than that in normal breast tissues, respectively (stage I: 94.8 (68.9-131.3), stage II: 96.0 (72.7-129.7), stage III: 98.3 (77.0-137.4), stage IV:105.7 (85.7-121.1), all P<0.001). The expression level of DCTPP1 in tissues of stage III breast cancer patients was significantly higher than that in stage I breast cancer tissues, and the difference was statistically significant (P<0.05).
Figure 2. Comparison of DCTPP1 mRNA Expression Levels between Normal Breast Tissues and Breast Lobular Carcinoma Tissues of Different Stages (A); normal breast tissues and breast lobular carcinoma tissues of different subclasses (B).
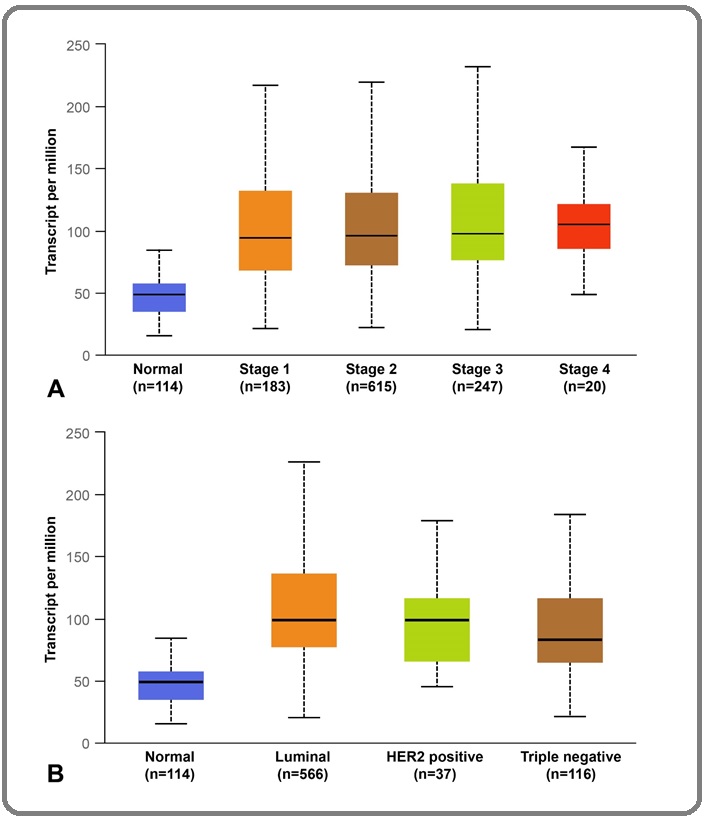
The expression level of DCTPP1 in tissue of patients with Luminal, HER-2 over-expression, and triple- negative breast lobular carcinoma was 98.7 (77.8-136.1) TPM, 99.0 (66.3-115.5) TPM, and 82.9 (65.3-115.8) TPM, repectively. The expression of DCTPP1 in each pathological subtype of breast cancer tissue was higher than that of normal breast tissue (all P<0.001). And the expression of this gene in Lumnial breast cancer was significantly higher than that in triple-negative breast cancer (P<0.001), as shown in Figure 2B.
Figure 2. Comparison of DCTPP1 mRNA expression levels between normal breast tissues and breast lobular carcinoma tissues of different stages (A); normal breast tissues and breast lobular carcinoma tissues of different subclasses (B).
The expression level of DCTPP1 protein between breast cancer tissues versus normal breast tissues
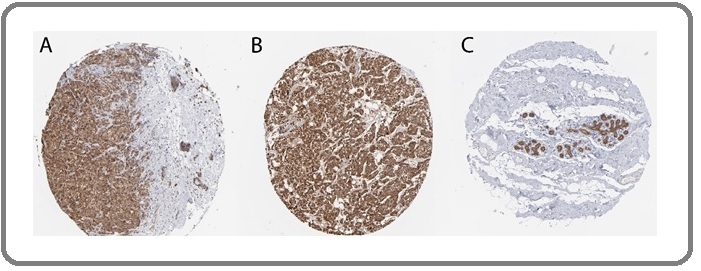
The results of immunohistochemistry in Figure 3A-B showed that the expression of DCTPP1 gene in breast lobular carcinoma tissue and breast ductal carcinoma tissue was strongly positive. Further observation of the distribution pattern of DCTPP1 protein-positive cells in breast cancer tissues revealed that the cell membrane, cytoplasm and nucleus of tumor cells are all expressed by DCTPP1 protein, among which the cytoplasm is light brown, and the nucleus and cell membrane are brown. In normal breast tissue, DCTPP1 is expressed in some gland cells, mainly located in the nucleus and cell membrane, and not expressed in adipose tissue, as shown in Figure 3C.
Figure 3. Immunohistochemical Determination of DCTPP1 Gene Expression in Breast Lobular Carcinoma Tissue (A), Breast Ductal Carcinoma Tissue (B), and Normal Breast Tissue (C).
The relationship between DCTPP1 expression level and the prognosis of breast cancer patients
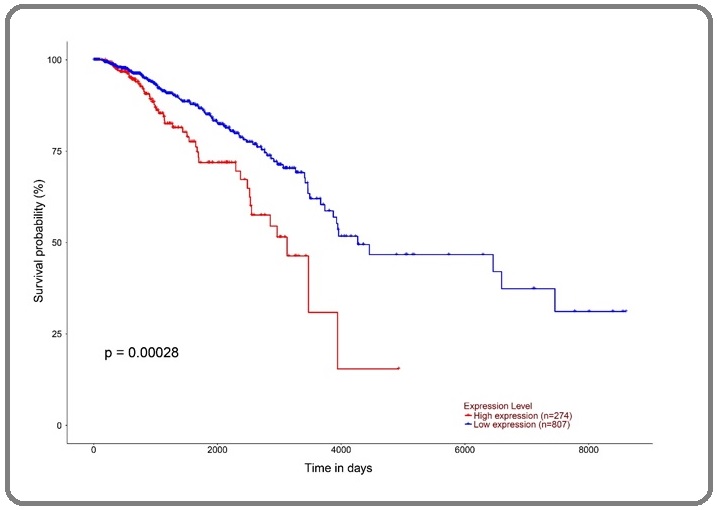
The TCGA database was used to analyze the association between the different expression levels of DCTPP1 gene and the prognosis of breast cancer patients, and the overall survival time of 1081 breast cancer patients was analyzed. The patients were divided into the high expression group (n=274) and the low expression group (n=807) based on the median of DCTPP1 gene expression, the follow up time was 25 years. Log-rank test showed that compared with low expression patients, the high expression of DCTPP1 gene reduced the overall survival time of breast cancer patients (HR=1.9, P<0.001), and the Kaplan-Meier curve was also showed in Figure 4.
Figure 4. Analysis of Overall Survival of Breast Cancer Patients with Different Expression Levels of DCTPP1 Gene Using TCGA Database.
DCTPP1 PPI network prediction and functional analysis
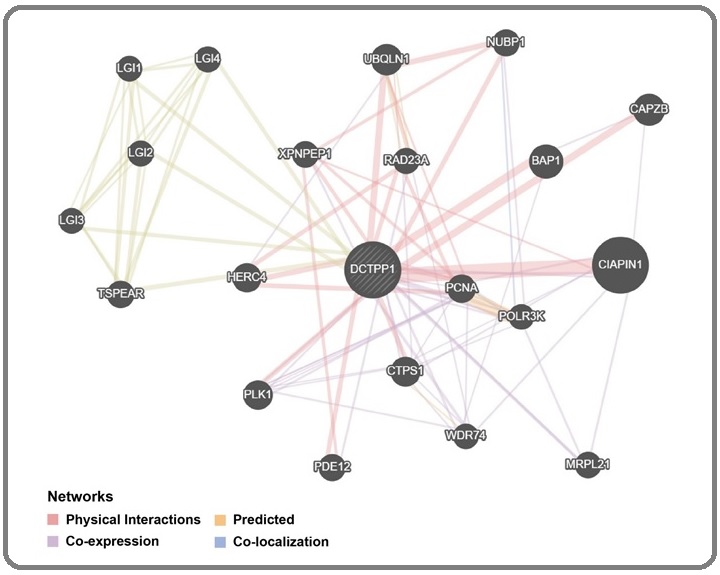
As shown in Figure 5, GeneMANIA analysis screened a total of 20 DCTPP1-interacting proteins, specifically including the following protein: Cytokine Induced Apoptosis Inhibitor 1 (CIAPIN1), BRCA1 Associated Protein 1 (BAP1), Capping Actin Protein Of Muscle Z-Line Subunit Beta (CAPZB), Cytidine Triphosphate Synthase 1 (CTPS1), HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 4 (HERC4), Leucine Rich Glioma Inactivated 1-4 (LGI1-4), Mitochondrial Ribosomal Protein L21 (MRPL21), Nucleotide Binding Protein 1 (NUBP1), Proliferating Cell Nuclear Antigen (PCNA), Phosphodiesterase 12 (PDE12), Polo Like Kinase 1 (PLK1), RNA Polymerase III Subunit K (POLR3K), RAD23 Homolog A Nucleotide Excision Repair Protein (RAD23A), Thrombospondin Type Laminin G Domain And EAR Repeats (TSPEAR), Ubiquilin 1 (UBQLN1), WD Repeat Domain 74 (WDR74), X-Prolyl Aminopeptidase 1 (XPNPEP1). The interaction relationship mainly included physical interaction, co-expression, and co-localization.
Figure 5. Protein-protein Interaction Analysis of DCTPP1 Gene.
GO analysis
Analyzing the biological process, cellular component, and molecular function of the above genes, it is found that these proteins are mainly involved in several processes such as biological regulation, response to stimulus, metabolic process, and cell communication (Figure 6A).
Figure 6. Analysis of Gene Ontology Associated with DCTPP1 Expression. A, Biological Process; B, Cellular Component; C, Molecular Function.
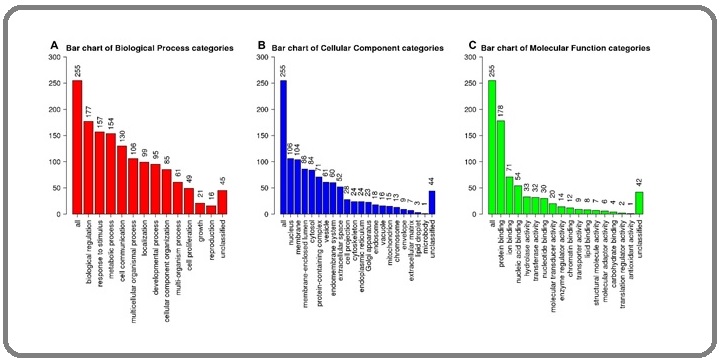
DCTPP1-related genes are located in a variety of cellular components, including nucleus, membrane, membrane- enclosed lumen, cytosol, protein-containing complex, vesicle, endomembrane system, etc (Figure 6B). Figure 6C indicated that its molecular functions included protein binding, ion binding, nucleic acid binding, hydrolase activity, transferase activity, nucleotide binding, etc.
Discussion
In recent years, the application of targeted drugs such as vascular endothelial growth factor, human epidermal growth factor receptor, cell cycle regulation by checkpoints, etc. in breast cancer has gradually increased. However, these targets are limited to some breast cancer patients. Therefore, searching for key molecules or targets for breast cancer is of great significance for the development of new targeted drugs for the treatment of breast cancer.
Normal cells in the human body are constantly undergoing self-renewal. One of the most critical steps is DNA replication, and this process is a very accurate process in the cell. Each gene produces 10-7 errors per generation [11]. Errors in this process can cause gene mutations, and some of the cells that undergo mutations will become precancerous cells. The precancerous cells transform into cancer cells under the action of the tumor microenvironment in the human body. Abnormal nucleotide metabolism in tumor cells plays a very important role in the formation and maintenance of tumor microenvironment, tumor cell proliferation, apoptosis and migration [12].
The NTP-PPase family exists in both prokaryotes and eukaryotes. The enzymes hydrolyze substrates including nucleoside triphosphates and abnormal nucleoside triphosphates [3]. Loss of NTP-PPase will lead to an increase in DNA mutations and affect cell division. For example, the MutT enzyme of E.coli can selectively remove the oxidized nucleotides 8-oxo-dGTP and 2-OH-dATP in the nucleotide pool. This process prevents the incorporation of abnormal nucleotides caused by oxidative stress into DNA, which can lead to DNA damage. The deletion of MutT results in a greatly increased mutation rate of AT-CG in E. coli [13]. MazG in Mycobacterium tuberculosis hydrolyzes dUTP, 2-hydroxy-dATP and 8-oxo-dGTP. The deletion of this enzyme can cause a significant increase in the mutation of the gene encoding rifampicin resistance, thereby affecting the normal growth of Mycobacterium tuberculosis [14]. Therefore, NTP-PPase continuously breaks down the abnormal nucleotides in the cell metabolism process in the biological process, ensuring the accuracy of DNA replication and the stability of the organism’s genome. As an NTP-PPase with a typical MazG domain found in human cells, the DCTPP1 gene is an active protein molecule with fewer studies and reports among many discovered NTP-Ppases. The results of its enzymatic properties and function studies suggest that the molecule can specifically hydrolyze dCTP and structural analogs such as 5-methyl-dCTP and 5-halo-DCTP [15].
The researchers studied the expression pattern of DCTPP1 gene in a variety of tumors and the expression difference between it and the tumor-adjacent tissues [5]. Compared with the expression in adjacent tissues, they found that DCTPP1 gene was significantly expressed in lung cancer tissues. There was no significant difference in the expression of DCTPP1 gene in gastric cancer and colorectal cancer, and the expression level was also different in tumors of different histological subtypes. The results of this study show that the expression of DCTPP1 gene in breast cancer tissue was significantly higher than that in normal breast tissue. This was the same as the expression pattern of the DCTPP1 gene in the above-mentioned tumor. The expression of DCTPP1 gene in different stages and different molecular pathological types (Luminal type, HER-2 positive type, triple-negative type) of breast cancer was higher than that of normal tissues, and it was more obvious in Lumnial type. In the Human Protein Atlas database, the expression of DCTPP1 protein in breast cancer tissue and normal breast tissue was detected by immunohistochemistry and we found that the expression of DCTPP1 protein in both breast ductal carcinoma and lobular carcinoma was higher than that in normal breast tissue. A research studied 161 cases of breast cancer tissues and 132 cases of adjacent tissues, and the results of immunohistochemistry showed that the DCTPP1 gene was overexpressed in cancer tissues and concentrated in the nucleus [15]. This result was consistent with the result of the present research, which was DCTPP1 gene is closely related to tumors.
After analyzing the overall survival of 1081 breast cancer patients, we found that the overall survival time of breast cancer patients with high expression of DCTPP1 was significantly shortened. Morisaki et al. [16] compared and analyzed the proteomics of gastric cancer stem cells and found that DCTPP1 gene activation may be related to DNA replication in gastric cancer stem cells, and they has been found that the high expression of DCTPP1 gene is related to the poor prognosis of gastric cancer patients. Lu et al. [17] also found the same in prostate cancer. These results suggested that the DCTPP1 gene may be a poor prognostic factor for cancer patients, cancer patients with high expression of DCTPP1 have a shorter survival time than patients with low expression of DCTPP1.
Requena et al. [18] explored the role of DCTPP1 gene in cell nucleotide homology, studied the extensive characterization of this enzyme, and found that the enzyme is ubiquitous in the nucleus, cytoplasm, and mitochondria. In normal cell metabolism, DCTPP1 can catalyze the hydrolysis of dCTP into dCMP, thereby maintaining the dCMP pool at the level of thymidylate synthesis. The proper ratio of dCTP/dTTP is very important for maintaining physiological homeostasis in the nucleotide pool. The activity of DCTPP1 gene in cells may be involved in the regulation of dCTP concentration in the nucleotide pool, and affect the ratio of dCTP/dTTP in the intracellular nucleotide pool. Existing studies have shown that NTP-PPase eliminates abnormal nucleotides in the process of cell metabolism and plays a clean-up function [3]. Xia et al. [4] subsequently found that the highly expressed DCTPP1 gene in gastric cancer promoted the resistance of gastric cancer to chemotherapy through the demethylation of MDR1. By further exploring the function of DCTPP1 gene through proteins related to DCTPP1 gene, it is found that such proteins are widely distributed in various structures of cells and participate in the metabolic growth process of cells and the process of protein binding. Lu et al. [17] found that over-expression of the DCTPP1 gene promoted the growth of prostate cancer tumor cells. Requena et al. [19] found that the DCTPP1 gene could increase the anti-tumor activity of chemotherapeutics in a study on the human cervical cancer cell line Hela.
In conclusion, the DCTPP1 gene was highly expressed in breast cancer tissues and is related to the poor prognosis of breast cancer patients. It may be involved in the occurrence and development of breast cancer. Further research on this gene can assist in evaluating the prognosis of patients and provide new intervention targets for the treatment of breast cancer.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for- profit sectors. The authors declare no conflict of interest.
References
- Cancer statistics, 2019 Siegel Rebecca L., Miller Kimberly D., Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2019;69(1). CrossRef
- Breast cancer statistics, 2013 DeSantis Carol, Ma Jiemin, Bryan Leah, Jemal Ahmedin. CA: A Cancer Journal for Clinicians.2013;64(1). CrossRef
- Crystal structure of the MazG-related nucleoside triphosphate pyrophosphohydrolase from Thermotoga maritima MSB8 Padmanabhan Balasundaram, Deshmukh Prashant, Yokoyama Shigeyuki, Bessho Yoshitaka. Journal of Structural and Functional Genomics.2015;16(2). CrossRef
- DCTPP1 attenuates the sensitivity of human gastric cancer cells to 5-fluorouracil by up-regulating MDR1 expression epigenetically Xia Li-liang, Tang Ya-bin, Song Fei-fei, Xu Ling, Ji Ping, Wang Shu-jun, Zhu Ji-min, Zhang Yong, Zhao Guo-ping, Wang Ying, Liu Tao-tao. Oncotarget.2016;7(42). CrossRef
- dCTP pyrophosphohydrase exhibits nucleic accumulation in multiple carcinomas Zhang Y., Ye W.Y., Wang J.Q., Wang S.J., Ji P., Zhou G.Y., Zhao G.P., Ge H.L., Wang Y.. European Journal of Histochemistry.2013;57(3). CrossRef
- DCTPP1 prevents a mutator phenotype through the modulation of dCTP, dTTP and dUTP pools Martínez-Arribas Blanca, Requena Cristina E., Pérez-Moreno Guiomar, Ruíz-Pérez Luis M., Vidal Antonio E., González-Pacanowska Dolores. Cellular and Molecular Life Sciences.2019;77(8). CrossRef
- UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses Chandrashekar Darshan S., Bashel Bhuwan, Balasubramanya Sai Akshaya Hodigere, Creighton Chad J., Ponce-Rodriguez Israel, Chakravarthi Balabhadrapatruni V.S.K., Varambally Sooryanarayana. Neoplasia.2017;19(8). CrossRef
- The human protein atlas: A spatial map of the human proteome Thul Peter J., Lindskog Cecilia. Protein Science.2017;27(1). CrossRef
- The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function Warde-Farley David, Donaldson Sylva L., Comes Ovi, Zuberi Khalid, Badrawi Rashad, Chao Pauline, Franz Max, Grouios Chris, Kazi Farzana, Lopes Christian Tannus, Maitland Anson, Mostafavi Sara, Montojo Jason, Shao Quentin, Wright George, Bader Gary D., Morris Quaid. Nucleic Acids Research.2010;38(suppl_2). CrossRef
- WebGestalt: an integrated system for exploring gene sets in various biological contexts Zhang B., Kirov S., Snoddy J.. Nucleic Acids Research.2005;33(Web Server). CrossRef
- Eukaryotic DNA Polymerases in Homologous Recombination McVey Mitch, Khodaverdian Varandt Y., Meyer Damon, Cerqueira Paula Gonçalves, Heyer Wolf-Dietrich. Annual Review of Genetics.2016;50(1). CrossRef
- Metabolic changes associated with tumor metastasis, part 2: Mitochondria, lipid and amino acid metabolism Porporato Paolo E., Payen Valéry L., Baselet Bjorn, Sonveaux Pierre. Cellular and Molecular Life Sciences.2015;73(7). CrossRef
- Nucleoside Diphosphate Kinase Escalates A-to-C Mutations in MutT-Deficient Strains of Escherichia coli Kapoor Indu, Emam Elhassan Ali Fathi, Shaw Abhirup, Varshney Umesh. Journal of Bacteriology.2019;202(1). CrossRef
- Chemical Genetics Reveals a Role of dCTP Pyrophosphatase 1 in Wnt Signaling Friese Alexandra, Kapoor Shobhna, Schneidewind Tabea, Vidadala Srinivasa Rao, Sardana Juhi, Brause Alexandra, Förster Tim, Bischoff Matthias, Wagner Jessica, Janning Petra, Ziegler Slava, Waldmann Herbert. Angewandte Chemie International Edition.2019;58(37). CrossRef
- Human dCTP pyrophosphatase 1 promotes breast cancer cell growth and stemness through the modulation on 5-methyl-dCTP metabolism and global hypomethylation Song F-f, Xia L-l, Ji P, Tang Y-b, Huang Z-m, Zhu L, Zhang J, Wang J-q, Zhao G-p, Ge H-l, Zhang Y, Wang Y. Oncogenesis.2015;4(6). CrossRef
- Comparative Proteomics Analysis of Gastric Cancer Stem Cells Morisaki Tamami, Yashiro Masakazu, Kakehashi Anna, Inagaki Azusa, Kinoshita Haruhito, Fukuoka Tatsunari, Kasashima Hiroaki, Masuda Go, Sakurai Katsunobu, Kubo Naoshi, Muguruma Kazuya, Ohira Masaichi, Wanibuchi Hideki, Hirakawa Kosei. PLoS ONE.2014;9(11). CrossRef
- Autophagy induced by overexpression of DCTPP1 promotes tumor progression and predicts poor clinical outcome in prostate cancer Lu Jianming, Dong Weimin, He Huichan, Han Zhaodong, Zhuo YangJia, Mo RuJun, Liang Yingke, Zhu JianGuo, Li Ruidong, Qu Han, Zhang Le, Wang Shibo, Ma Renyuan, Jia Zhenyu, Zhong Weide. International Journal of Biological Macromolecules.2018;118. CrossRef
- The NTP pyrophosphatase DCTPP1 contributes to the homoeostasis and cleansing of the dNTP pool in human cells Requena Cristina E., Pérez-Moreno Guiomar, Ruiz-Pérez Luis M., Vidal Antonio E., González-Pacanowska Dolores. Biochemical Journal.2014;459(1). CrossRef
- The nucleotidohydrolases DCTPP1 and dUTPase are involved in the cellular response to decitabine Requena Cristina E., Pérez-Moreno Guiomar, Horváth András, Vértessy Beáta G., Ruiz-Pérez Luis M., González-Pacanowska Dolores, Vidal Antonio E.. Biochemical Journal.2016;473(17). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details