MDM2 (T309G) Gene Polymorphism Determines the Susceptibility of Hepatocellular Carcinoma in Bangladesh
Download
Abstract
Background: Hepatocellular carcinoma (HCC) is one of the fatal cancer types worldwide, and a variety of genetic factors are considered to be associated with this incidence. MDM2 gene plays a pivotal role in various pathways, which are essential to combat tumor formation. The study aimed to find out the associations of MDM2 (T309G, rs2279744) gene polymorphism with the development of HCC in the Bangladeshi population.
Methods: A case-control study on 100 HCC patients and 110 control subjects was conducted. The genotyping of the MDM2 (T309G) gene was done using PCR-RFLP methods.
Results: The percentage of TT and GG genotypes were significantly different (p<0.01) among the study subjects. There were four genotyping groups, while the subjects with TT genotypes were considered the reference group. Patients with GG genotypes were at high risk of developing HCC (OR, 3.6; 95 % CI, 1.64–7.80; p<0.01) compared to the control. On the other hand, the association of TG genotypes with HCC was not statistically significant (OR, 1.8; 95 % CI, 0.91–3.40, p>0.05). In addition, patients having either GG or TG genotypes showed higher risk for HCC compared to control group (OR = 2.20; 95% CI = 1.21–4.14; P < 0.05).
Conclusion: Our study suggested that the MDM2 gene may have a strong association with the development of HCC, and the GG allele could serve as an essential determinant to identify the higher risk of HCC in the Bangladeshi population.
Introduction
Hepatocellular carcinoma (HCC) is considered the fifth most common cancer worldwide, leading to over 600,000 deaths each year [1]. There are multiple risk factors, including chronic HBV or HCV infection, cirrhosis, excessive alcohol consumption, carcinogen exposure, and various genetic factors associated with HCC [2-5]. Genetic factors are accounted as inherited polymorphisms in the genes involved in cell-cycle regulation, DNA repair, and apoptosis [6, 7]. Hepatocarcinogenic risk factors could be defined by the identification of polymorphism of modifier genes. Murine double minute 2 (MDM2) plays a vital role not only in the p53 pathway but also in various other pathways such as inhibition of retinoblastoma protein (RB) and p73 as well as stabilization of p63 [8-12]. The MDM2 protein level is positively regulated by p53, while MDM2 negatively mediates p53 and activity through an auto-regulatory feedback loop [13-15]. In addition, MDM2 acts as an oncogene and plays a regulatory role by interacting with other tumor-related genes, which are inevitable for cell-cycle control.
The genomic size of the human MDM2 gene is 34 kb with two promoters, the p53-responsive intronic promoter and the constitutive promoter [16]. Although MDM2 is rarely mutated, it is highly polymorphic. A naturally occurring functional SNP (rs2279744) in the p53-response intronic promoter of the MDM2 gene has recently been described by Bond et al. (2004), which is referred to as SNP309 or T309G [17]. The exchange of T to G at 309 position of the p53-responsive intronic promoter augments the binding affinity of the Sp1 transcription activator, causing the increased level of MDM2, which directly inhibits p53 transcriptional activity facilitating the transcription of the damaged cells to escape from the cell-cycle checkpoint and become carcinogenic [17, 18]. The MDM2 polymorphism has been associated with almost all forms of cancer. Several studies have reported that the MDM2 (T309G) polymorphism is associated with susceptibility to various tumors, including gastric carcinoma [19], non-small-cell lung cancer [20], endometrial cancer [21], sarcoma [17], and bladder cancer [22]. Some recent investigations have shown the association of MDM2 (T309G) polymorphism with the risk of HCC in Chinese [23], Japanese [24], Korean [25], and Turkish [26]. On the other hand, contradictory results were reported where no association between MDM2 (T309G) polymorphism and HCC had been shown in different populations [27, 28].
The study of underlying mechanisms of complex diseases such as cancer by identifying SNPs that alter the expression and function of genes contributing to the predisposition of diseases has become a prominent area of investigation. Since the interaction of MDM2 and TP53 plays pivotal roles in apoptotic cell death, DNA repair, and cell-cycle checkpoint, we hypothesized that the genetic polymorphism in MDM2 might contribute to the susceptibility to hepatocellular carcinogenesis. The MDM2 (T309G) gene polymorphism and its relationship with the susceptibility of HCC may vary in different populations to different ethnic groups, depending on different geographical areas and levels of environmental exposures. To the best of our knowledge, an association between MDM2 (T309G) polymorphism and HCC susceptibility has not yet been studied in the Bangladeshi population. The present study aimed to determine the effects of MDM2 (T309G) polymorphism on the risk of developing hepatocellular carcinoma in Bangladeshi people.
Materials and Methods
This study was conducted on 210 subjects, including 100 patients suffering from hepatocellular carcinoma and a total of 110 healthy control subjects (Table 1).
| Healthy Control | HCC Patients (n=100, %) | |
| (n=110, %) | ||
| Age | 47.5 ± 13 | 51.3 ± 16 |
| Gender | ||
| Male | 89 (81) | 78 (78) |
| Female | 21 (19) | 22 (22) |
| Alcohol intake | ||
| Yes | 02 (1.8) | 02 (2) |
| No | 108 (98.2) | 98 (98) |
| Family history of cancer | ||
| Yes | - | 15 (15) |
| No | - | 65 (65) |
| No comments | - | 20 (20) |
Results are Expressed as Number (percentage) and Mean±SD
We found no significant differences in baseline characteristics among the study subjects (Table 1). The HCC cancer patients who had no history of other chronic diseases were recruited from the Department of Hepatology of Bangabandhu Sheikh Mujib Medical University, Dhaka. The healthy controls subjects who don’t have any history of cancer or chronic diseases were assigned in this study during their regular health check-up in different hospitals of Dhaka city (Table 1).
The nature of the study was explained among the participants, and informed consent from them was obtained. A structured questionnaire covering the information of age, gender, medical and family history of chronic diseases was completed. The ethical review committee (ERC) of the Department of Biochemistry and Molecular Biology, University of Dhaka, approved the study. In this study, the rules and regulations of the declaration of Helsinki and its subsequent revisions were followed [29].
Sample collection
All aseptic precautions were taken, and disposable syringes were used to draw approximately 3.0 mL of venous blood from each individual with the help of a trained phlebotomist. EDTA (1.20 mg/mL) containing tubes were used to collect the drawn blood and transferred immediately into the laboratory in an icebox and stored at −20°C until genomic DNA extraction.
Genotyping of MDM2 (T309G) Gene
The MDM2 (T309G) genotypes were determined using our previously described PCR-RFLP method [30]. Bailes’s method was followed to extract the genomic DNA from peripheral leukocytes [31]. Then the genomic DNA was amplified by polymerase chain reaction (PCR). PCR conditions and primer sequences were used according to the method of Walsh et al. (2007) [21]. The Go Taq polymerase from Promega Corporation, USA, was used to carry out PCR. Approximately 0.5 μg of genomic DNA was added to a PCR mix which was composed of 200 μmol dNTPs, 50 pmol of each primer, 2.5 units Taq polymerase, and PCR buffer consisting of 10 mol/mL Tris-HCl (pH 8.3), 2.5 mol/mL MgCl2 and 50 mol/mL KCl in a volume of 50 μL.
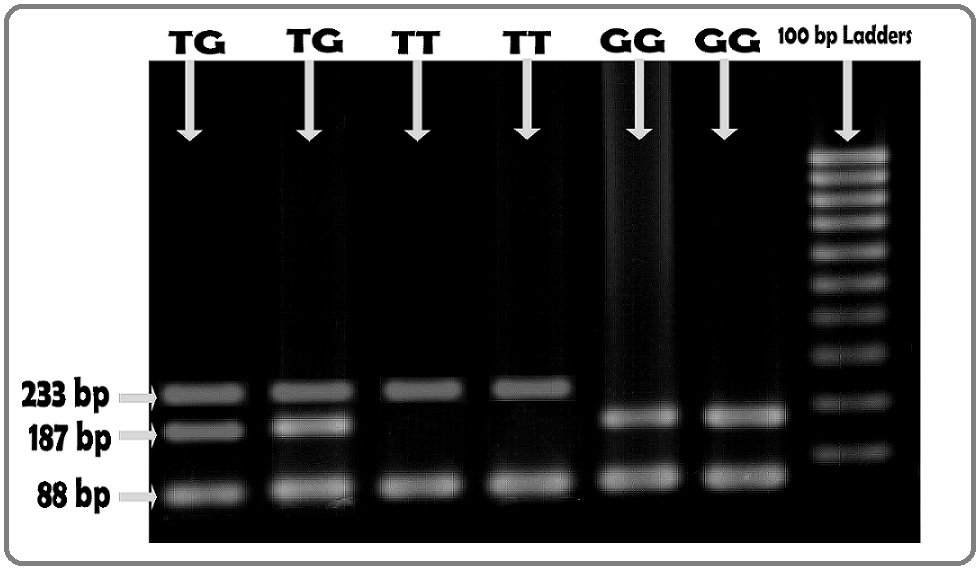
About 10.0 µL PCR product (352 bp) for T309G genotype was digested with MspA1I restriction enzyme (New England Biolabs, Ipswich, MA, USA) at 37oC for 30-60 min. The T/T, T/G, and G/G genotypes were identified as 233/88 bp, 233/187/88 bp, and 187/88 bp, respectively, running on the 3% agarose gel electrophoresis and the 46 bp of the complete digestion of 233 bp fragment could not be resolved (Figure 1).
Figure 1. Banding Pattern of MDM2 (T309G) Gene in 3% Agarose Gel. Presence of 233/187/88 bd fragments, 233/88 bp fragments and 187/88 bp fragments illustrated the presence of TG, TT and GG genotypes respectively. Because of small size, the 46 bp band of complete digestion of 233 bp fragment could not be resolved.
Statistical Analysis
The Statistical Package for Social Science (SPSS), windows version 17.0, was used to perform statistical analysis. The odds ratios (ORs) were calculated to assess the relative association between cases and controls. As a measure of relative risk, ORs at 95% confidence intervals (95% CI) were estimated using logistic regression models. The results were considered statistically significant when the p-value is <0.05.
Results
This study examined the association of MDM2 (T309G) gene polymorphism as a risk factor for developing hepatocellular carcinoma. The baseline characteristics showed no significant difference between the two groups (Table 1). The age and gender between the control and cases have been matched. Moreover, the alcohol consumption data and family history showed no association with HCC in the Bangladeshi population. This is may be due to the law prohibiting alcohol consumption in Bangladesh. The genotypic analysis was done and presented in Table 2 and Table 3.
| Genotypes | Controls subjects | HCC patients | p value |
| (n=110, %) | (n=100, %) | ||
| TT | 41 (37.3) | 21 (21) | <0.01 |
| TG | 51 (46.4) | 46 (46) | |
| GG | 18 (16.3) | 33 (33) |
Results are expressed as number (percentage). Chi-square test was performed. p<0.05 was taken as level of significance.
| Genotypes | Control (N=110) | Patients (N=100) | p value | OR (95%, CI) |
| TT | 41 | 21 | - | 1 (Ref.) |
| TG | 51 | 46 | ns | 1.8 (0.91 – 3.40) |
| GG | 18 | 33 | <0.01 | 3.6 (1.64 – 7.80) |
| GG+TG | 69 | 79 | <0.05 | 2.2 (1.21 – 4.14) |
Results expressed as number. Fisher’s exact test was done to evaluate significance among genotypic groups. 95% CI; 95% confidence interval; p< 0.05 was taken as level of significance; ns, not significant.
Genotypic distribution and analysis of MDM2 (T309G) genotypes
Table 2 represents the genotype frequency of the MDM2 (T309G) gene in different study groups. The TT and GG genotypes were significantly different (p<0.01) between the study subjects. The percentages of TT, TG, and GG alleles were 37.3%, 46.4%, and 18.3%, respectively, in control subjects. On the other hand, the percentages of TT, TG, and GG alleles were 21%, 46%, and 33%, respectively, in HCC patients.
The risk of developing HCC associated with the MDM2 (T309G) genotypes was estimated (Table 3). There were four genotyping groups, while the subjects with TT genotypes were considered the reference group. Patients with GG genotypes were at high risk of developing HCC (OR, 3.6; 95 % CI, 1.64–7.80; p<0.01) compared to the control. On the other hand, an association of TG genotypes with HCC was not statistically significant (OR, 1.8; 95 % CI, 0.91–3.40, p>0.05). In addition, patients having either GG or TG genotypes showed higher risk for HCC compared to control group (OR = 2.20; 95% CI = 1.21–4.14; p< 0.05).
Discussion
Our present study was a molecular epidemiologic case-control study where we examined whether the MDM2 (T309G) polymorphisms influence the risk of HCC in the Bangladeshi population. The T309G SNP (rs2279744) is situated in the intronic promoter region of the MDM2 gene, and our recent study revealed that T309G SNP was associated with the increased risk of HCC in the Bangladeshi population. This finding is consistent with previous studies where the T309G allele has been reported as the risk allele for HCC [23, 24, 32], and also suggests that the MDM2 T309G polymorphism may be used as one of the biomarkers for genetic susceptibility to HCC. The association of MDM2 T309G polymorphism with hepatocellular carcinoma susceptibility has been shown previously [17, 18, 23, 24, 27, 28]. In recent studies by Qiu et al. (2016) and Akkiz et al. (2010) have shown the association of MDM2 T309G polymorphism with HCC susceptibility [25, 26]. Our results were also consistent with the results of Japanese [23], Moroccan [32], Korean [24], and Chinese populations [27]. On the other hand, Leu et al. (2009) found no association between the MDM2 T309G genotypes and HCC susceptibility in the Taiwanese population [28]. The apparent lack of association of the study conducted by Leu et al. (2009) may result from a low sample size as well as unmatched age and gender of the study population [28]. In addition, the contribution of genetic polymorphisms to the risk for cancer may depend on environmental factors, geographic and ethnic differences.
As MDM2 has been evidenced to interact with several key tumor suppressors, including RB [11] and p53 [16], MDM2 polymorphism might affect cancer risk in several ways. First of all, MDM2 is a vital regulator of the TP53 pathway that facilitates the degradation of TP53, and over expression or augmentation of MDM2 has been frequently reported in almost all forms of human cancer types [33]. Secondly, the over expression of MDM2 has been associated with the susceptibility to carcinogenesis in genetically modified animals. Jones et al. (1998) showed that the MDM2-transgenic mice produce four-fold more MDM2 developed spontaneous tumors while transgenic mice with normal MDM2 levels did not develop tumors [34]. Thirdly, the binding affinity of Sp1 to the promoter of MDM2 is augmented by the G allele at 309 position ofMDM2 gene, increasing the cellular activity of MDM2 that abolished TP53 DNA damage response [17]. Therefore, considering the role of MDM2 in cancer formation, one might assume that individuals carrying the G allele may have a high level of MDM2 over a lifetime and would be at higher risk for developing hepatocellular carcinoma and other cancer.
In conclusion, this study provides evidence that polymorphism in MDM2 is associated with the risk of developing hepatocellular cancer in the Bangladeshi population. The G allele of MDM2 T309G could serve as an important marker to identify a higher risk of HCC. These results support the hypothesis that naturally occurring genetic variation in the tumor suppressor TP53 pathway may associate with cancer susceptibility. However, as this is the first report concerning the MDM2 polymorphism and the risk of HCC in the Bangladeshi population, independent studies with a large sample size are needed to validate our findings.
Acknowledgments
We would like to acknowledge the support and cooperation of the participants of this study. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors reported that there is no conflict of interest.
References
- Global Cancer Statistics, 2002 Parkin D. M., Bray F., Ferlay J., Pisani P.. CA: A Cancer Journal for Clinicians.2005;55(2). CrossRef
- Hepatocellular carcinoma pathogenesis: from genes to environment Farazi Paraskevi A., DePinho Ronald A.. Nature Reviews Cancer.2006;6(9). CrossRef
- Molecular Pathogenesis of Hepatocellular Carcinoma McKillop Iain H., Moran Diarmuid M., Jin Xiaoling, Koniaris Leonidas G.. Journal of Surgical Research.2006;136(1). CrossRef
- Molecular pathogenesis of human hepatocellular carcinoma Thorgeirsson Snorri S., Grisham Joe W.. Nature Genetics.2002;31(4). CrossRef
- Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis El–Serag Hashem B., Rudolph K. Lenhard. Gastroenterology.2007;132(7). CrossRef
- Hepatocellular Carcinoma and Polymorphisms in Carcinogen-Metabolizing and DNA Repair Enzymes in a Population with Aflatoxin Exposure and Hepatitis B Virus Endemicity Kirk G. D.. Cancer Epidemiology Biomarkers & Prevention.2005;14(2). CrossRef
- Putative Association of Fas and FasL Gene Polymorphisms with Clinical Outcomes of Hepatitis B Virus Infection Jung Yong Jin, Kim Yoon Jun, Kim Lyoung Hyo, Lee Soo Ok, Park Byung Lae, Shin Hyoung Doo, Lee Hyo-Suk. Intervirology.2007;50(5). CrossRef
- The PTEN, Mdm2, p53 tumor suppressor–oncoprotein network Mayo Lindsey D, Donner David B. Trends in Biochemical Sciences.2002;27(9). CrossRef
- The p53 pathway: positive and negative feedback loops Harris Sandra L, Levine Arnold J. Oncogene.2005;24(17). CrossRef
- Mdm2 binds p73α without targeting degradation Bálint Eva, Bates Stewart, Vousden Karen H. Oncogene.1999;18(27). CrossRef
- Interaction between the retinoblastoma protein and the oncoprotein MDM2 Xiao Zhi-Xlong, Chen Jiandong, Levine Arnold J., Modjtahedi Nazanlne, Xing Jun, Sellers William R., Livingston David M.. Nature.1995;375(6533). CrossRef
- The Human MDM2 Oncoprotein Increases the Transcriptional Activity and the Protein Level of the p53 Homolog p63 Calabrò Viola, Mansueto Gelsomina, Parisi Tiziana, Vivo Maria, Calogero Raffaele A., La Mantia Girolama. Journal of Biological Chemistry.2002;277(4). CrossRef
- Functions of the MDM2 oncoprotein Freedman D. A., Wu L., Levine A. J.. Cellular and Molecular Life Sciences CMLS.1999;55(1). CrossRef
- Moll UM and Petrenko O (2003). The MDM2-p53 interaction. Mol Cancer Res, 1, 1001-8. .
- Mdm2 promotes the rapid degradation of p53 Haupt Ygal, Maya Ruth, Kazaz Anat, Oren Moshe. Nature.1997;387(6630). CrossRef
- Momand J and Zambetti GP (1997). Mdm-2: ‘big brother’ of p53. J Cell Biochem, 64, 343-52. .
- A Single Nucleotide Polymorphism in the MDM2 Promoter Attenuates the p53 Tumor Suppressor Pathway and Accelerates Tumor Formation in Humans Bond Gareth L., Hu Wenwei, Bond Elisabeth E., Robins Harlan, Lutzker Stuart G., Arva Nicoleta C., Bargonetti Jill, Bartel Frank, Taubert Helge, Wuerl Peter, Onel Kenan, Yip Linwah, Hwang Shih-Jen, Strong Louise C., Lozano Guillermina, Levine Arnold J.. Cell.2004;119(5). CrossRef
- Genetic polymorphisms in cell cycle regulatory genesMDM2 andTP53 are associated with susceptibility to lung cancer Zhang Xuemei, Miao Xiaoping, Guo Yongli, Tan Wen, Zhou Yifeng, Sun Tong, Wang Yonggang, Lin Dongxin. Human Mutation.2005;27(1). CrossRef
- MDM2Promoter Polymorphism Is Associated With Both an Increased Susceptibility to Gastric Carcinoma and Poor Prognosis Ohmiya Naoki, Taguchi Ayumu, Mabuchi Nobuyuki, Itoh Akihiro, Hirooka Yoshiki, Niwa Yasumasa, Goto Hidemi. Journal of Clinical Oncology.2006;24(27). CrossRef
- Association of a functional polymorphism in the promoter of theMDM2 gene with risk of nonsmall cell lung cancer Lind Helge, Zienolddiny Shanbeh, Ekstrøm Per Olav, Skaug Vidar, Haugen Aage. International Journal of Cancer.2006;119(3). CrossRef
- Association between a functional single nucleotide polymorphism in the MDM2 gene and sporadic endometrial cancer risk Walsh Christine S., Miller Carl W., Karlan Beth Y., Koeffler H. Phillip. Gynecologic Oncology.2007;104(3). CrossRef
- Onat OE, Tez M, Ozcelik T et al (2006). MDM2 T309G polymorphism is associated with bladder cancer. Anticancer Res, 26, 3473–5. .
- MDM2 Promoter SNP309 Is Associated with the Risk of Hepatocellular Carcinoma in Patients with Chronic Hepatitis C Dharel Narayan, Kato Naoya, Muroyama Ryosuke, Moriyama Masaru, Shao Run-Xuan, Kawabe Takao, Omata Masao. Clinical Cancer Research.2006;12(16). CrossRef
- MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection Yoon Y. J., Chang H. Y., Ahn S. H., Kim J. K., Park Y. K., Kang D. R., Park J. Y., Myoung S. M., Kim D. Y., Chon C. Y., Han K.-H.. Carcinogenesis.2008;29(6). CrossRef
- Interaction between p53 codon 72 and MDM2 309T>G polymorphisms and the risk of hepatocellular carcinoma Qiu Moqin, Liu Yingchun, Yu Xiangyuan, Qin Linyuan, Bei Chunhua, Zeng Xiaoyun, Qiu Xiaoqiang, Tang Bo, He Songqing, Yu Hongping. Tumor Biology.2015;37(3). CrossRef
- MDM2 promoter polymorphism is associated with increased susceptibility to hepatocellular carcinoma in Turkish population Akkız Hikmet, Sümbül Ahmet Taner, Bayram Süleyman, Bekar Aynur, Akgöllü Ersin. Cancer Epidemiology.2010;34(4). CrossRef
- Jiang D (2008). p53 gene mutation, p53 R72P and MDM2 SNP309 polymorphisms and hepatocellular carcinoma development and prognosis [D]. Fudan University, 121-3. .
- Association between MDM2-SNP309 and hepatocellularcarcinoma in Taiwanese population Leu Jyh-Der, Lin I-Feng, Sun Ying-Fang, Chen Su-Mei, Liu Chih-Chao, Lee Yi-Jang. World Journal of Gastroenterology.2009;15(44). CrossRef
- World Medical Association Declaration of Helsinki JAMA.2013;310(20). CrossRef
- Association of TP53 gene polymorphisms with susceptibility of bladder cancer in Bangladeshi population Hosen Md. Bayejid, Salam Md. Abdus, Islam Md. Fakhrul, Hossain Ashfaque, Hawlader M Zakir Hossain, Kabir Yearul. Tumor Biology.2015;36(8). CrossRef
- An Inexpensive, Simple Protocol for DNA Isolation from Blood for High-Throughput Genotyping by Polymerase Chain Reaction or Restriction Endonuclease Digestion Bailes S.M., Devers J.J., Kirby J.D., Rhoads D.D.. Poultry Science.2007;86(1). CrossRef
- MDM2 SNP309T>G polymorphism and risk of hepatocellular carcinoma: A case–control analysis in a Moroccan population Ezzikouri Sayeh, El Feydi Abdellah Essaid, Afifi Rajae, El Kihal Latifa, Benazzouz Mustapha, Hassar Mohammed, Marchio Agnès, Pineau Pascal, Benjelloun Soumaya. Cancer Detection and Prevention.2009;32(5-6). CrossRef
- Amplification of a gene encoding a p53-associated protein in human sarcomas Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B.. Nature.1992;358(6381). CrossRef
- Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis Jones S. N., Hancock A. R., Vogel H., Donehower L. A., Bradley A.. Proceedings of the National Academy of Sciences.1998;95(26). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details