Alu-deletion in Bangladeshi Women: Scopes of Clinical Utility for the Detection of Early Breast Cancer as a Biomarker
Download
Abstract
Objective: Alteration of Alu elements of human DNA may result cancer. This study was aimed to find status of Alu-deletion among breast cancer patients.
Methods: To identify Alu-deletion, tissue samples from the operated specimen and blood of the breast cancer patients were collected. For comparison, blood was collected from healthy controls. After extraction, DNA was amplified by PCR with Alu specific primer and gel electrophoresis was done. Alu specific banding patterns of DNA of the samples were examined and compared.
Result: Among the 64 patients and equal number (64) of healthy women examined, multiple Alu polymorphic loci have been identified. One of them characterized as deletion at the 2000bp region, found in 83% cancerous blood samples and only 3% in the healthy controls (p < 0.0001), OR 149. Another band was identified as less intensified at the 1700bp, present in 74% cancerous blood samples (p < 0.0001). We also found differential DNA banding pattern between breast cancer (7-9) and control blood samples (9-15) which is highly significant (p < 0.0001). Whereas no such banding pattern was observed in cancer tissues and normal breast tissues collected during surgery.
Conclusion: This study confirms significant presence of Alu-deletion in women with breast cancer in comparison without cancer. Alu-deletion can be presumed as a useful blood-based biomarker for the detection of early breast cancer.
Introduction
Human cancer undoubtedly recognized now-a-days as genetic disease. From the day of David Hahnemann (1858–1920) to present days scientist reached to a consensus that genes affect pathways of cell growth, cell signaling, apoptosis and Deoxyribonucleic Acid (DNA) repair which finally lead to oncogenesis [1, 2].
There are small deletions and insertions which are described as point mutations in different descriptions of genetic disorders. Besides these, chromosomal deletions and loss of heterozygosity are responsible for activation of tumor suppressor genes. Insertion or loss of a transoposon element in the genome dynamics was amenable for DNA damage interfering with gene activities. It may lead to different genetic diseases including cancer [3, 4].
They are also considered as non-allelic homologous recombination factor causing copy number variation and different disease [5, 6]. As a part of that, Alu-element came to the center of discussion which may explain the origin of some diseases including breast cancer [7]. Alu elements exist as a group of DNA sequences called inserted repetitive DNA. Some literatures describe Alu-repeat as the most important and rich repeats in the human genome. They apparently lack a general function. The enzyme Alu-I influences the DNA base pair for its existence. So they are named after the enzyme Alu-I [8, 9].
Alu-elements are sequences of nucleotides which was familiar as SINEs (short interspersed nuclear elements. They account for 10% of human genome [8]. Alu-elements are inherited from the 7SL RNA (Ribonucleic acid) gene [2] [4].
They make a broad family of mobile genetic elements within the human genome. Through retroposition they can duplicate themselves via an RNA intermediate theory and increase their number. They are approximately 300 base pair long in sequence and mostly obtained from the introns, untranslated regions of genes and intergenic genomic regions [8]. They are described as the most common transposable elements that induce insertion mutagenesis [10]. Insertions and deletions of them are detectable by PCR(Polymerase chain reaction).
Scientist predicted that over more than 60 million years Alu elements multiply to more than one million copies among the primate genomes. Accordingly, they produced a group of subfamilies with time. During gene expression Alu elements influence the genome and gene activity by insertion mutation, gene conversion, deletion [8]. Alu elements modify the gene function by Alu insertion into the exons. Like Alu- insertion, Alu-deletion events also occur in human genome by Alu insertion-mediated deletions (AIMD) or Alu recombination-mediated deletions (ARMD). Genomic rearrangements by Alu insertion cause about 0.1% of human diseases and genomic deletions lead to ARMD for 0.3% of human genetic disorders [11, 12]. It is also descried that there are many Alu elements which may cause many human diseases [13] .
Researchers predicted that during the process of Alu amplification there may be recombination between dispersed Alu elements which might result gene changes including duplications, deletions and insertions. All may lead to genomic instability resulting initiation of different disease processes including cancer [14].
In the era of personalized medicine, there are tremendous development on genetics. Many evidences prevails expressing multiple genes are responsible for the development of hereditary or non-hereditary breast cancer. Sometimes genetic tests are guiding to subgroup the disease into several types for the purposes of diagnostic and therapeutic options. No single gene is yet to be identified as key matchmaker of the process, rather it is a common impression that breast cancer is heterogeneous disease. Accordingly different molecular kits are now available to stratify the disease for treatment and prognostic subtyping.
National Comprehensive Cancer Network [15] and other large bodies in the field of breast cancer management in the world are yet to find the definite guideline and molecular component for screening and early detection before the appearance of symptoms [16].
This study was aimed and designed to find out the status of Alu-deletion in Bangladeshi women including breast cancer patients by using Alu-PCR acquiring fingerprints of cancer and normal tissues. The finding was also matched with the clinicopathological findings of the breast cancer patients like age, tumor status, hormone receptor status, and family history of cancer to get any relationship with the altered Alu-polymorphism picture.
Materials and Methods
Case selection and sample collection
This study was undertaken by an understanding of collaboration between Centre for Advanced Research in Sciences (CARS), Dhaka University and Department of surgical oncology of National institute of cancer research and hospital, Dhaka, Bangladesh between 2015-18. Patients takes their services in the hospital across the country. The work was approved by the ethical committee, University of Dhaka. Candidates were selected from the admitted patients for surgical treatment after the diagnosis of invasive ductal carcinoma of breast. Samples were taken from the patients in the operation day once or twice a month. Four to six samples were collected in a single day, who were underwent for breast cancer surgery. Cases were selected randomly as both the patients and investigators were blind about their identity and profile. Tumor and normal tissue samples were taken from the operated specimen by the surgical team. Blood samples were taken before the procedure from the same patients. A healthy cohort of equal number of females matching their age, ethnicity, socioeconomic status from the mainstream population residing in Bangladesh were included. Blood (B), breast cancer tissues (CT), and approximately 2.3 cm2 peripheral normal tissues (NT) were obtained from all breast cancer patients during surgery. Blood samples from healthy individuals were considered as control females (CF). Breast cancer patients were interviewed and detailed informations were taken from their record. Informed written consent was taken from each patient for inclusion in the study.
About 5 mL of blood samples were collected in EDTA coated tubes from the selected breast cancer patients and control samples after proper counseling. The tissue samples were transported to the laboratory at 4oC cool box and kept at -20°C until DNA was extracted.
DNA isolation, Alu-PCR and Gel Electrophoresis
DNA from the blood samples was isolated by standard proteinase K treatment thereafter with phenol/chloroform/ isoamyl alcohol extraction. Besides, DNA from tissue samples was extracted by the above mentioned method. DNA was precipitated by mixing with the solution of 0.3 M sodium acetate (pH 5.2) in 70% ethanol at -20°C overnight treatment and resuspended in Tris-EDTA (TE) at buffer (pH 8). The quantification of DNA was performed by taking absorbance at 260 nm and visualized by 0.8% agarose gel electrophoresis. DNA isolated from tumor tissue, normal tissue of the operated breast and blood was amplified by PCR. DNA between Alu-1 repeats was amplified with the use of up to three different Alu 1 primers. The Alu specific three primers were used. The Alu specific three primers were (R12A/267: AGCGAGACTCCG; R14B/264: CAGAGCGAGACTCT; and A/8: TGAGCCACCGCG) [17]. To wind up our result, one primer A/8 was selected finally. The 50 μL of reactions were performed in 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 100 mM dNTPs each 1 µM primer, 1 U of TaqMan™ DNA polymerase (Applied Biosystem, USA) and 250 ng DNA template. The PCR amplification response was guided by following thermal conditions: initial denaturing at 94°C for 7 min, 27 amplification cycle (30 s at 94°C, 45s at 50°C and 120 s at 72°C), and extension at 72°C for 7 min. Samples were amplified until the DNA fingerprint was firmly validated. Following amplification, PCR products were run in parallel in adjacent lanes during electrophoresis to separate DNA fragments according to length. The resulting DNA fragments were visualized as a series of bands on nondenaturing 1.5% agarose with ethidium bromide nucleic acid staining.
Statistical Analysis
Data were analyzed in GraphPad Prism® v 8, (GraphPad Software Inc., La Jolla, CA) to verify the Alu change probability difference in cancer and healthy (control) populations [18]. To confirm the result of this common test in the light of low expected number in the healthy population Yates’s chi and un-corrected chi squared test (‘N-1’ chi squared test) have been used as expected to give relatively low Type I error, significance was taken ≤.05. The methods used to compute (confidence interval) Cls with Koopmans asymptotic score, (odd ratio) OR with Baptista-Pike, sensitivity, and specificity etc. with Wilson-Brown [19].
Results
In total, 64 patients could be examined, taking the same number (64) of healthy control subjects. Table 1 summarizes patients’ profile, it showed that mean age was 49.7 years and controls’ mean age was 45.8 years.
| Variables | Number | Percentage | ||
| Parameters | Subgroups | |||
| < 40 | 19 | 29.6 | ||
| Controls (n=64) | ≥ 40 | 45 | 70.3 | |
| Mean | 45.8 | |||
| Age in years | < 40 | 18 | 28.1 | |
| Patients (n=64) | ≥ 40 | 46 | 71.8 | |
| Mean | 49.7 | |||
| Positive family history of cancer of patients | Yes | 8 | 12.5 | |
| No | 56 | 87.5 | ||
| Tumor Stage | T1+T2 | 42 | 65.6 | |
| T3+T4 | 22 | 34.3 | ||
| Gr I | 24 | 37.5 | ||
| Tumor grade | Gr II | 30 | 46.8 | |
| Gr III | 10 | 15.6 | ||
| HR Positive | 28 | 43.7 | ||
| Hormonal status | HR Negative | 36 | 56.2 | |
| Yes | 38 | 59.3 | ||
| Chemotherapy status | No | 26 | 40.6 |
Positive family history of cancer in the patient group was found in 12%, no information was available in the control group. Patients with early breast cancer were over 65%. Immunochemical examination showed that nearly 44 percent patients were hormone positive (HR+ve) and rest were hormone –ve cancers. Nearly 60% patient received neoadjuvant chemotherapy.
From the gel electrophoresis screening, differences in the number of DNA bands between control and breast cancer samples were obtained ranging from 300bp to 2000bp. These bands were compared for understanding the number of bands appeared and for band intensity. Among 64 control blood samples, sixty-two (62) samples showed no changes in their DNA bands shown in Figure 1.
Figure 1. Comparison of Cancer Blood Samples and Control Blood Samples in their DNA Bands.
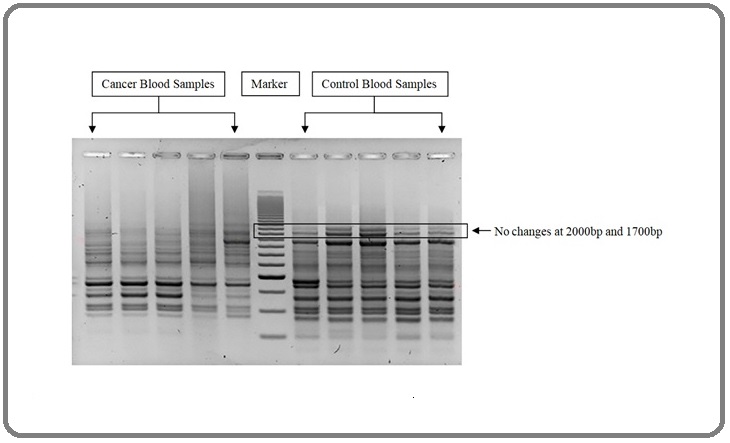
The cancer blood samples showed complete deletion (83%) of bands at 2000bp (p < 0.0001), OR (149.4), CI (95%) 31.13 to 649.2 described in Table 2.
| Subject | Fragment size (bp) | Control (n = 64) | Breast cancer (n = 64) | P value | OR | CI (95%) |
| Alu Deletion | 2000 | 2.23 % (2) | 82.81 % (53) | <0.0001 | 149.4 | 31.13 to 649.2 |
| Alu Low intensity | 1700 | 73.44 % (47) | 7.81 % (5) | <0.0001 | 32.62 | 11.18 to 82.35 |
| Alu band number | 100-2000 | 8.188 | 11.92 | <0.0001 | - | 3.194 to 4.275 |
Sensitivity for Alu-deletion was 82.81% and specificity 96.88% with positive predictive value (PPV) 96.36% and negative predictive value (NPV) 84.93%. Whereas less intense bands (74%) at 1700bp (p < 0.0001) OR (32.62), CI (95%) 11.18 to 82.35 with sensitivity 71.79%, specificity 89.30% with PPV 87.68% and NPV 75,00%. Similar Alu bands deletion was observed in 68% breast cancer tissues but absent in peripheral tissues shown in Figure 2a and Figure 2b.
Figure 2. a, Changes at 1700bp and 2000bp in Cancer Blood Samples; b. Comparison of Cancer Tissue Samples with Peripheral Tissue and Control Blood Samples. (Only cancer tissue samples show absence of bands at 2000bp and 1700bp).
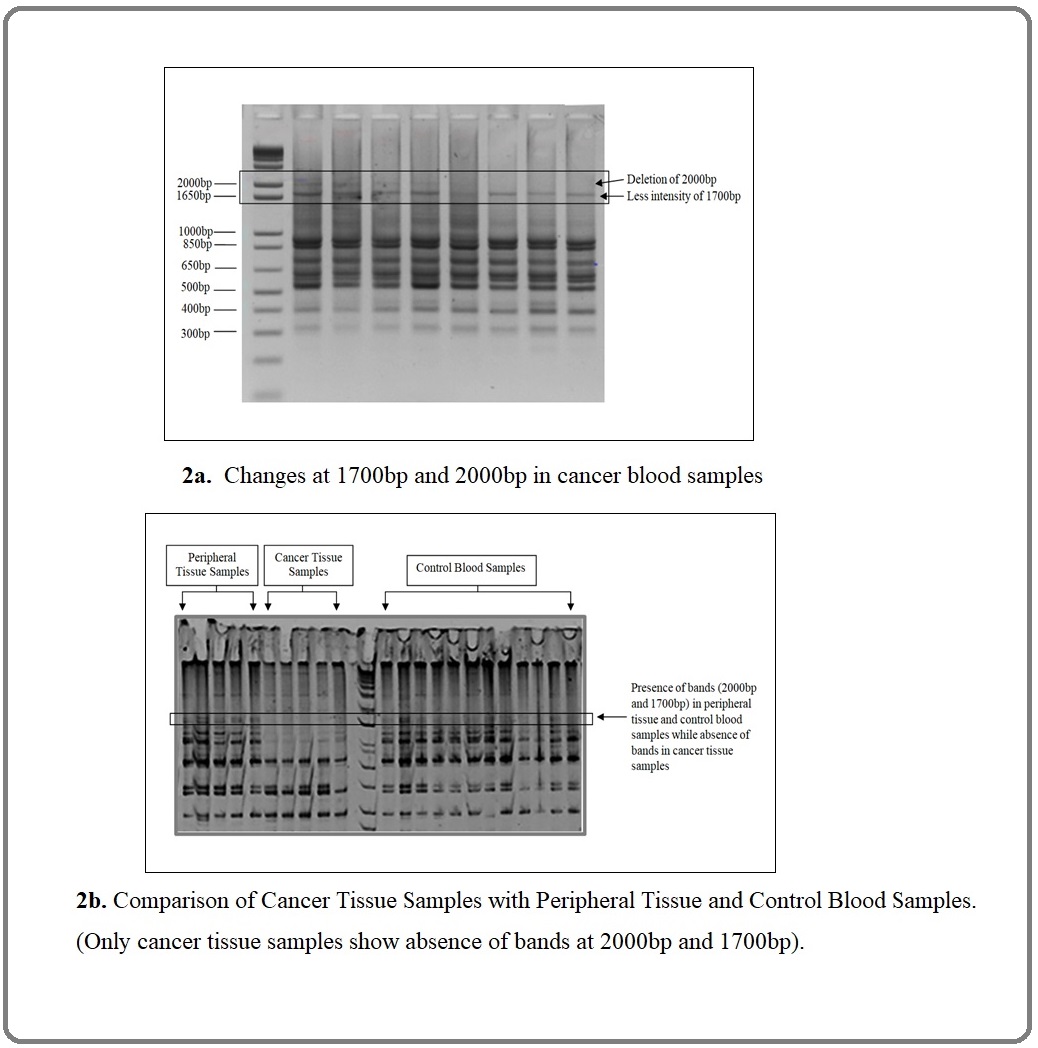
We compared the number of bands in both groups (Table 2) showing that in control blood samples which vary from 9-15 whereas in cancer blood samples, the number of bands vary from 7 to 9 only (p < 0.0001), CI (3.19 to 4.27) displayed in Figure 3.
Figure 3. Differential Banding Pattern in Control and Breast Cancer Blood Samples.

Changes in the number of bands expressed irrespective of tumor stage, grade and hormonal status and age of the patients.
Discussion
Breast cancer marker commonly depends on the specific gene expression [20]. Breast cancer marker e.g. carcinoembryonic antigen (CEA), and cancer antigen (CA15-3) showed some efficacy in diagnosing metastatic breast cancer but they too can’t monitor breast cancer in preliminary steps [21]. For early detection of breast cancer, there are ongoing investigations on some potential serum and apoptotic biomarkers [22] but still, there is lacking in the development of sensitive blood-based biomarkers [23]. In an attempt to increase marker sensitivity and specificity, an approach has been focused based on the blood-based Alu-PCR technique to investigate the insertion and/or deletion of Alu elements in breast cancer comparing to healthy control.
In the current case control study, we selected 64 breast cancer patients to identify their Alu- banding status in the human genome after DNA extraction from the tissue taken from tumor area and non-tumor area. 64 Blood sample was also taken from the patients to make a comparison with the healthy control subjects.
Though family history is a modest factor, it has a pivotal role in the development of breast cancer [24]. In our study, most cancer patients (76.56%) did not have any family history of breast cancer. Hence, other factors could be involved like age, nutrition, physical activity, etc. In a study, it has been suggested that half of the Bangladeshi breast cancer patients are ER and PR positive and two in every five cases are Her2/neu over expressed [25]. In the case of hormonal status, our study suggests that breast cancer in Bangladesh is related to hormone receptor gene expression.
Breast cancer DNA samples were represented as Alu banding pattern by gel electrophoresis technique. It was observed that in breast cancer patients, blood samples had significant complete deletion of Alu 2000bp band (83%) in comparison to equal number of healthy controls, (97%) which showed no changes in their DNA bands. Thus it is a customary thinking most of the cancer patients might have Alu-deletion in their DNA repeats.
There is limited study in the literature on Alu-deletion, but few studies founds on nucleotide of the DNA polymorphism called INDEL which means insertion or deletion of bases in the genome [25]. The proportion of INDEL varies in different population of the world. Study found that short length INDEL or Alu have the capacity of deletion: insertion by 4:1. This value is consistent with Human Gene Mutation Database [26]. One study in India suggested from unrelated individuals, Alu elements were found to be highly polymorphic. Their coefficient of gene differentiation was higher than populations in most other global regions, except Africa [27].
There are numbers of studies on Alu insertion [28,29]. On the other hand only, few studies reveal that Alu-mediated delusion also occurs in the human genome, but numbers are less in comparison with the insertions in different studies. The deletion incidences occur through Alu-insertion mediated deletions (AIMD) or Alu recombination-mediated deletions (ARMD). There are chances of deletions during interchromosomal recombination depending on the arrangement of DNA sequences. The current study reveals similar results of Alu deletion in cancer patients which might be due to rearrangement of DNA sequences.
Pauline et al. [30] in their study quantified the role of genomic Alu retro transposition-mediated deletion (ARD) to the instability of the primate genome, the ARD rate in humans is about 0.21%. But other study showed much higher frequencies of ARD of between 0.8% and 8% [31] . It is a general concept that Alu repeats play an important role in genome evolution and some cellular processes [8][32]. All are associated with genetic instability that may be counted as a principal event in oncogenesis. Alu-mediated mutagenesis by insertion and recombination have been reported in a few cancer genes [9][33]. In cancer biology, genetic instability was assumed as one of the key factors for the development of cancer [11]. Study describes that Alu elements are presumed to be a leading factor to cause human genomic instability resulting different human diseases. Fazza A.C. et al. [7] in their work on breast cancer patients’ genomic DNA from tumor tissue and normal breast tissue, tissue from surgical margin and from blood sample analyzed with Alu-PCR. They reveled average loss of one band for every 3.3 altered bands. Surgical margins also disclose altered Alu-PCR profile with frequencies of 0.5-0.9.
It is a settled issue that early diagnosis and treatment gives a remarkable outcome in the treatment of breast cancer. Data reveals that survival for early-stage (stage 1) breast cancer is nearly 100% after 5 years from diagnosis. On the contrary survival for metastatic breast cancer (stage 4) reduced to 32% at 5 years from diagnosis [34]. Thus to catch the disease at an early stage now a strong objective for the caregivers across the globe to control the disease. The most intriguing finding from our study is the difference in the number of bands between cancer and control blood samples. Consequently, it can be explained that the sequence between two Alu elements may be more susceptible to deletion and/or unequal recombination to form a new arrangement. Through the Alu-PCR technique, the deletion of band thus provides a fingerprinting profile of quantitative and qualitative changes in the sample studied here. Therefore, the change in number of the bands and deletion of 2000bp band can be conducive as a preliminary strategy for identifying regions of genomic instability involved in the initiation and progression of breast cancer.
For early detection, measures like clinical examination and self-examination in the low-income countries and mammographical screening are existing protocol in the affluent countries.
Though debates exist regarding benefit of mammography in the international arena considering chances of overdiagnosis, overtreatment and hazards of radiation [35]. In addition, mammograms cannot detect smaller non palpable lesions and are less accurate in cancer detection in women with dense breasts. Therefore, in future days it will be necessary to develop alternative tools for early detection of breast cancer [36]. So, attention is given for other tools particularly towards the use of biomarkers.
There are many reports of using non-invasive body fluid-based tests [37]. For example, circulating carcinoma antigens, circulating tumor cells, circulating microRNAs and other tests in the peripheral blood, nipple aspirate fluid or from other body fluids [36, 38]. But each component has got its diversity considering diagnostic or prognostic use, sensitivity, and specificity. Alu-deletion could be a newer tool to consider for early detection of breast cancer. In our present work, its sensitivity and specificity of Alu deletion of the breast cancer patients was 83% and positive predictively was 97%, so from the point of cancer diagnosis, finding was much favorable in comparison with control, it is seen that OR of the Alu-deletion result was also remarkable. Alu-loss was also seen among the different age group, tumor group, hormone status and tumor stages. So, we can predict deletion can be seen even in early stages of the disease even in the phase of nonpalpable stage. So, it can be a useful component to detect early breast cancer particularly for screening purposes, high risk patients.
In conclusion, Genomic sequencing studies reveal that distribution of uniform Alu elements are required for genomic stability. Alu-deletion may be one of the key oncologic components for genomic instability resulting breast cancer in the study population. In the healthy population this aberration is rarely available. So, it can be presumed to be an useful and potential blood-based biomarker for the diagnosis of breast cancer even in early stage. It may be an appropriate tool to detect the non-palpable cancer, as a result there are scopes to put forward new potentiality for screening breast cancer in the population.
Acknowledgements
Thanks to Ministry of Education, Government of Bangladesh for their support with fund. We are also thankful to Professor Zeba Islam Seraj for supporting us with PAGE experiment facilities.
Conflict of Interest
The authors declare no conflicts of interest.
Limitations
Alarger sample size would have been produced a better convincing result.
References
- Cancer genes and the pathways they control Vogelstein Bert, Kinzler Kenneth W.. Nature Medicine.2004;10(8). CrossRef
- Epigenetic changes in cancer Iacobuzio-Donahue Christine A.. Annual Review of Pathology.2009;4. CrossRef
- Alu sequences are processed 7SL RNA genes Ullu E., Tschudi C.. Nature.1984;312(5990). CrossRef
- The Mobile Genetic Element "Alu" in the Human Genome Novick GE, Batzer MA, Deininger PL, Herrera RJ. Bio Science.1996;46(1):32-41.
- Widespread establishment and regulatory impact of Alu exons in human genes Shen Shihao, Lin Lan, Cai James J., Jiang Peng, Kenkel Elizabeth J., Stroik Mallory R., Sato Seiko, Davidson Beverly L., Xing Yi. Proceedings of the National Academy of Sciences of the United States of America.2011;108(7). CrossRef
- Alu elements: know the SINEs Deininger Prescott. Genome biology.2011;12. CrossRef
- Estimating genomic instability mediated by Alu retroelements in breast cancer Fazza Ana Cristina, Sabino Flavia Cal, Setta Nathalia, Bordin Newton Antonio, Silva Eloiza Helena Tajara, Carareto Claudia Marcia Aparecida. Genetics and Molecular Biology.2009;32(1). CrossRef
- Alu repeats and human genomic diversity Batzer Mark A., Deininger Prescott L.. Nature Reviews. Genetics.2002;3(5). CrossRef
- Alu elements of the human genome Andreassen R. Tidsskr Nor Laegeforen.2004;124(18):2345-2349.
- Transposable Elements in Human Cancer: Causes and Consequences of Deregulation Anwar Sumadi Lukman, Wulaningsih Wahyu, Lehmann Ulrich. International Journal of Molecular Sciences.2017;18(5). CrossRef
- Alu repeats and human disease Deininger P. L., Batzer M. A.. Molecular Genetics and Metabolism.1999;67(3). CrossRef
- Alu retrotransposition-mediated deletion Callinan Pauline A., Wang Jianxin, Herke Scott W., Garber Randall K., Liang Ping, Batzer Mark A.. Journal of Molecular Biology.2005;348(4). CrossRef
- Structural Variation of Alu Element and Human Disease Kim Songmi, Cho Chun-Sung, Han Kyudong, Lee Jungnam. Genomics & Informatics.2016;14(3). CrossRef
- Alu Mobile Elements: From Junk DNA to Genomic Gems Dridi Sami. Scientifica.2012;2012. CrossRef
- National Comprehensive Cancer Network(NCCN). Genetic/familial high-risk assessment for: breast and ovarian (Version 2.2019)..
- Genomics applied to the treatment of breast cancer Hamdan Diaddin, Nguyen Thi Thuy, Leboeuf Christophe, Meles Solveig, Janin Anne, Bousquet Guilhem. Oncotarget.2019;10(46). CrossRef
- DNA fingerprints provide a patient-specific breast cancer marker Toth-Fejel SuEllen, Muller Patrick, Ham Bruce, Esvelt Kevin, Dumas Nicole, Calhoun Kristine, Pommier Rodney. Annals of Surgical Oncology.2004;11(6). CrossRef
- GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, California USA,www.graphpad.com .
- "Probable inference, the law of succession and statistical inference". Wilson E. B. J Am Med Assos.2012;22:209-212. JSTOR 2276774.
- Molecular markers for breast cancer: prediction on tumor behavior Banin Hirata Bruna Karina, Oda Julie Massayo Maeda, Losi Guembarovski Roberta, Ariza Carolina Batista, Oliveira Carlos Eduardo Coral, Watanabe Maria Angelica Ehara. Disease Markers.2014;2014. CrossRef
- Circulating cell free DNA as blood based biomarker in breast cancer Stötzer OJ, Lehner J, Braun M, Holdenrieder S. Mol Biol.2014;3:120.
- Prediction of response to neoadjuvant chemotherapy in breast cancer patients by circulating apoptotic biomarkers nucleosomes, DNAse, cytokeratin-18 fragments and survivin Stoetzer OJ, Fersching DMI, Salat C, Steinkohl O , et al . Cancer Lett.2013;336(1):140-148.
- National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers Sturgeon Catharine M., Duffy Michael J., Stenman Ulf-Håkan, Lilja Hans, Brünner Nils, Chan Daniel W., Babaian Richard, Bast Robert C., Dowell Barry, Esteva Francisco J., Haglund Caj, Harbeck Nadia, Hayes Daniel F., Holten-Andersen Mads, Klee George G., Lamerz Rolf, Looijenga Leendert H., Molina Rafael, Nielsen Hans Jørgen, Rittenhouse Harry, Semjonow Axel, Shih Ie-Ming, Sibley Paul, Sölétormos György, Stephan Carsten, Sokoll Lori, Hoffman Barry R., Diamandis Eleftherios P.. Clinical Chemistry.2008;54(12). CrossRef
- Family history and risk of breast cancer: an analysis accounting for family structure Brewer Hannah R., Jones Michael E., Schoemaker Minouk J., Ashworth Alan, Swerdlow Anthony J.. Breast Cancer Research and Treatment.2017;165(1). CrossRef
- Evolution of hormone receptor status in breast carcinoma Begum KNA, Akond Ak, Huq N, Aymon NN, Huq F. J Shaheed Suhrawardy. Med. Coll.2018;10(02):70-72.
- Spontaneous microdeletions and microinsertions in a transgenic mouse mutation detection system: analysis of age, tissue, and sequence specificity Halangoda A., Still J. G., Hill K. A., Sommer S. S.. Environmental and Molecular Mutagenesis.2001;37(4). CrossRef
- Human-specific insertion/deletion polymorphisms in Indian populations and their possible evolutionary implications Majumder P. P., Roy B., Banerjee S., Chakraborty M., Dey B., Mukherjee N., Roy M., Thakurta P. G., Sil S. K.. European journal of human genetics: EJHG.1999;7(4). CrossRef
- Transposable Elements: No More 'Junk DNA' Kim Yun-Ji, Lee Jungnam, Han Kyudong. Genomics & Informatics.2012;10(4). CrossRef
- Recently mobilized transposons in the human and chimpanzee genomes Mills Ryan E., Bennett E. Andrew, Iskow Rebecca C., Luttig Christopher T., Tsui Circe, Pittard W. Stephen, Devine Scott E.. American Journal of Human Genetics.2006;78(4). CrossRef
- Alu retrotransposition-mediated deletion Callinan Pauline A., Wang Jianxin, Herke Scott W., Garber Randall K., Liang Ping, Batzer Mark A.. Journal of Molecular Biology.2005;348(4). CrossRef
- Recently integrated Alu elements and human genomic diversity Salem Abdel-Halim, Kilroy Gail E., Watkins W. Scott, Jorde Lynn B., Batzer Mark A.. Molecular Biology and Evolution.2003;20(8). CrossRef
- Comparative Analysis of transposed element insertion within human and mouse genes reveals Alu's unique role in shaping the human transcriptome Sela Noa, Mersch Britta, Gal-Mark Nurit, Lev-Maor Galit, Hotz-Wagenblatt Agnes, Ast Gil. Genome biology.2007;8. CrossRef
- Alu Elements Deininger P. 2006. CrossRef
- Relative Survival by Stage at Diagnosis (Female Breast Cancer) Available online: https://ncci.canceraustralia.gov.au/relative-survival-stage-diagnosis-female-breast-cancer (accessed 3 May 2021)..
- Benefits and harms of mammography screening Løberg Magnus, Lousdal Mette Lise, Bretthauer Michael, Kalager Mette. Breast Cancer Research : BCR.2015;17(1). CrossRef
- Why the Gold Standard Approach by Mammography Demands Extension by Multiomics? Application of Liquid Biopsy miRNA Profiles to Breast Cancer Disease Management Zubor Pavol, Kubatka Peter, Kajo Karol, Dankova Zuzana, Polacek Hubert, Bielik Tibor, Kudela Erik, Samec Marek, Liskova Alena, Vlcakova Dominika, Kulkovska Tatiana, Stastny Igor, Holubekova Veronika, Bujnak Jan, Laucekova Zuzana, Büsselberg Dietrich, Adamek Mariusz, Kuhn Walther, Danko Jan, Golubnitschaja Olga. International Journal of Molecular Sciences.2019;20(12). CrossRef
- The future of blood-based biomarkers for the early detection of breast cancer Loke Sau Yeen, Lee Ann Siew Gek. European Journal of Cancer (Oxford, England: 1990).2018;92. CrossRef
- Liquid biopsy in breast cancer: A comprehensive review Alimirzaie Sahar, Bagherzadeh Maryam, Akbari Mohammad R.. Clinical Genetics.2019;95(6). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details