The Role of Autophagy-related Proteins of Beclin-1/BECN1, LC3II, and p62/SQSTM1 in Melanoma Tumors
Download
Abstract
Purpose: The prognosis of melanoma depends on early diagnosis and timely treatment. Autophagy plays a dual role in tumor progression. In the early stages, it prevents tumor formation, while in the advanced stages it promotes tumorigenicity. This study aimed at investigating the role of autophagy in different stages of melanoma and evaluating the relationship between autophagy and clinicopathological factors.
Methods: ATG5 and BECN1 genes expression in melanoma tumors were evaluated in a retrospective study of 5 years in the cancer institute of Tehran, Iran. The autophagy-related proteins and the correlation with clinicopathological data were also investigated in a tissue microarray series of 52 melanoma tumors following by immunohistochemical staining for the autophagy-associated proteins p62/SQSTM1 (p62), LC3II and Beclin-1/BECN1. The possibility of autophagy biomarkers was also predicted by ROC curve analysis.
Results: ATG5 and BECN1 gene expression decreased in melanoma tumors in comparison with tumor margins. However, BECN1 expression at the protein level increased with tumor progression. The expression of LC3II also raised while the p62 level declined as the tumor progressed, suggesting an increased autophagy activity during tumor development. Furthermore, melanoma ulceration was positively correlated with BECN1, LC3II and p62 expression with p<0.05, though the melanoma mitotic rate and thickness did not significantly correlate with autophagy–related proteins expression.
Conclusions: Autophagy-related proteins are suggested as potential prognostic factors in melanoma and could be considered as a therapeutic target.
Introduction
Melanoma is one of the most dangerous types of skin cancer caused by the abnormal proliferation of melanocytes (pigment cells) [1]. The incidence of melanoma is increasing globally each year, which raised the mortality rate [2]. To reduce melanoma-related mortality and well prognosis, effectual diagnostic biomarkers are always under investigation [3,4]. It has demonstrated that the expression levels of many autophagy-related genes and proteins altered during melanoma malignancies, which might be considered as biomarkers for predicting its stages, invasiveness, patient survival, and treatment targets [5]. Macroautophagy (hereinafter referred to as autophagy) is a central lysosome-dependent mechanism activated to digest or reuse cellular contents in critical conditions such as nutrient deficiency and pathogenic infection to keep cell homeostasis [6]. Indeed, the contents of cell cytoplasm engulfs inside a specialized vacuole, called the autophagosome, which fuses with the lysosome to degrade the materials [7]. In this regulated process, the essential proteins are involved and named the AuTophaGy-related (ATG). Up to now, at least 41 ATG genes have been recognized during the autophagy process [8]. Among autophagy proteins, ATG5 is essential for autophagosome formation. Knocking down of ATG5 leads to downregulation of autophagy inhibition, suggesting that ATG5 plays a central role in autophagy [8,9]. Beclin-1/ BECN1 is a mammalian protein that is the ortholog of the yeast autophagy-related gene 6 (Atg6) and is involved in both the autophagy (initial step of autophagosome formation) and cell death signaling pathway by interacting to either phosphoinositol-3 kinase (PI3k) class III or BCL-2 [10].
LC3 (microtubule-associated protein 1A/1B-light chain 3) comprises a soluble LC3 I and a lipidated form called LC3 II. LC3 II is recruited into autophagosomes [11]. Different stressors promote the conjugation of LC3 I to phosphatidylethanolamine to organize the autophagosome-specific LC3 II, which is considered the most reliable marker of autophagy [12].
p62/SQSTM1 (p62) is an adaptor protein transporting ubiquitinated proteins during autophagy [13] and plays a substantial role in selective autophagic degradation of several cargoes. Generally, p62 and its substrate break down within the autophagolysosome. This process mediates by interaction with LC3, which is involved in phagophore/isolation membrane and autophagosome formation [14]. Impaired autophagy during tumorigenesis leads to p62/ubiquitinated protein aggregates [14] and visualizes as autophagy marker [13].
The function of autophagy in cancer is quite complex and somewhat controversial. In the early stages of cancer onset, autophagy activated as tumor-suppressive to prevent precancerous cells’ proliferation. However, in the later stages, it supports tumor stability by increasing the resistance of cancer cells to chemotherapy and or radiotherapy; consequently, inhibiting or activating autophagy in different stages of various cancers is a significant concern in treatment [6,15]. Since previous studies have shown the diagnostic and therapeutic importance of the autophagy pathway in melanoma, this study aimed to investigate the critical mediators involved in the autophagy pathway (ATG5, BECN1, p62, and LC3II) in different stages of melanoma and investigate whether autophagy is associated with clinopathological factors.
Materials and Methods
Patient samples
All the tumors of melanoma from 2015 to 2020 were separated and selected from tumor bank of Imam Khomeini Hospital, at Cancer Institute of Tehran, Iran. According to confirmation of pathologist, fifty two formalin-fixed paraffin-embedded (FFPE) tissue tumors and 52 tumor margin samples as controls were included in this study. The research was performed after approval of the local ethical committee of Iran with R.TUMS.VCR.REC.1398.076 ethical code (https:// ethics.research.ac.ir/EthicsProposalView.php?id=59775). The clinicopathological data were listed related to each patient’s pathology sheets. Regarding CAP approved of 2020 (College of American Pathologists protocol for the examination of excision specimens from patients with melanoma of the skin), all the available pathology features were recorded as follows; ulceration (present or absent), thickness (pTX: primary tumor, the thickness cannot be assessed, pT0: No evidence of primary tumor, pT1: 1.0 mm or less in thickness, pT2: tumor >1.0 to 2.0 mm in thickness, pT3: tumor>2.0 to 4.0 mm in thickness, pT4: tumor>4.0 mm in thickness), mitotic rate, lymph node metastasis (present and absent), distant metastasis (present and absent) and staging (anatomic or Clark level-based I for intra epidermal tumor only, II for tumor present in but does not fill and expand papillary dermis, III for tumor fills and expands papillary dermis, IV for tumor invades into reticular dermis, V for tumor invades subcutis).
RNA extraction
The RNA extraction from FFPE tissues was carried out manually according to our lab protocol. Briefly, the tumor portion of five shaved cuts (11 μm thickness) from each selected block was deparaffinized with xylene. Then they were centrifuged at high speed and washed several times with 70% ethanol. The pellet was kept at room temperature until the ethanol evaporated. A centrifuge was performed in each step, followed by adding lysis buffer, chloroform, cold isopropanol, and 70% ethanol. The supernatant was then drained and allowed to dry completely. Finally, the extracted RNA was kept in DEPC water, and the concentration was assessed using a NanoDrop™ OneC spectrophotometer (Thermo Fisher Scientific Inc). Optical density (OD) of samples was measured at 260 nm and 280 nm, and the ratio of 260/280 was used to test for protein or phenol contamination.
cDNA synthesis and qPCR
1000 ng / µl RNA was transferred to cDNA using the BIOFACT kit (South Korea, cat.no:BR631-096) according to the company’s instructions. The mRNA expression levels were measured by qPCR using specific pairs of primers in the following program: initial denaturation (10 min for 95 °C), denaturation (10 s for 95 °C), annealing (30 s for 60 °C, 40 cycles), and elongation (10 s for 72 °C). The oligonucleotide sequences of primers are as follow ATG5 F 5′-CACAAGCAACTCTGGATGGGATTG-3′ and ATG5 R 5′-GCCACAGGACGAAACAGCTTC-3′. BECN1 F 5′- AGGTTGAGAAAGGCGAGACA-3′, BECN1 R 5′-AATTGTGAGGACACCCAAGC-3′. GPDH F 5′-AATCCC ATCACCATCTTCCA-3′ and GPDH R:
5′-TGGACTCCACGACATACTCA-3′. The fold change of each gene was calculated by the 2 -ΔΔCt formula, where ΔΔCt=ΔCt target-ΔCt Contorol[16].
Biomarker evaluation
One of the standard methods for predicting the possibility of gene biomarkers is using the Receiver Operating Characteristics (ROC) curve. This graph shows the sensitivity against specificity across a range of cut points for a biomarker. The area under the curve (AUC) varies between 0.50 and 1.0, while 0.5 displays no discrimination between cases and controls, 1.0 indicates the best distinction. This would be present the probability of a case being accepted as a biomarker is 50% versus 100% [17].
Tissue microarray
To characterize immunohistochemical protein expression, three-block arrays were constructed for three different autophagy biomarkers. Briefly, two 1.5-mm-diameter cylinders of tissue were taken from representative areas of each archival paraffin block and arrayed into a new recipient paraffin block. In addition, standard margins of tumors and normal tissues were placed as internal controls to ensure the quality of staining slides.
Immunohistochemistry
Three-micrometer tissue sections from the TMA blocks were sectioned and applied to unique immunohistochemistry coated slides. These slides were baked overnight in a 56°C oven. Then they were deparaffinized by xylene, rehydrated through a graded ethanol series, and washed with phosphate-buffered saline. A 2-minute heat treatment achieved antigen retrieval in a pressure cooker containing 1 liter of 10 mmol/L sodium citrate buffer that had been previously brought to the boil. Following by quenching the endogenous peroxidase activity with 1.5% hydrogen peroxide in methanol for 10 minutes, Immunohistochemically staining was accomplished on these sections using three different antibodies, (LC3β (H-50): sc-28266, p62/ SQSTM1 (D-3): sc-28359 and BECN1 (E-8) sc-48341, all were from Santa Cruz Biotechnology, Inc. Hematoxylin and Eosin staining (H&E) was carried out on these selected tissues to anticipate the staining intensities using a standard protocol [18].
Interpretation of tissue immunohistochemical reactions
The IHC sample scoring system is a method for converting qualitative data into measurable and comparable data. Scoring is based on the intensity of coloring and the number of positive staining of the markers in the tumor cells, as follows; the amount of coloring cells or dot-like is scored as 0 for no staining or <5% of the cells visible, score 1 means weak staining for 5-25% of the cells detectable, score 2 means intermediate staining for 25-75% of the cells staining and score 3 means strong staining for > 75% of cells staining visible. The intensity staining score is also performed as follows; none for no cells visible, score 1 for < 1% of the cells detectable, score 2 for 1-10% of the cells staining, score 3 for 11-33% of the cells staining, score 4 for 34-66% of the cells staining and score 5 for 67-100% of staining. Furthermore, the normal tissues according to the data sheet were stained with the same autophagic markers and revealed null or very low expression. To correlate the pathological data with the immunohistochemical scores, they were classified as low (for scores o and 1) and high (for scores 2 and 3) [19].
Statistics
All data are from three independent experimentsand reported as the mean±standarddeviation (SD). The correlation of clinical data and immunohistochemistry scoring were performed using SPSS22 software, Ver19, with Chi-squared, Fisher exact, and Pearson Chi-Square tests. In all experiments, p≤0.05 was considered statistically significant. Data of biomarkers was illustrated with ROC curve using Graph pad Prism 8 software.
Results
Clinical data
The series comprises 52 patients diagnosed with cutaneous malignant melanoma. They were 24 females (46.2%) and 28 males (53.8%). Their ages at the time of diagnosis ranged from 25 to 87 years (mean, 56 years). According to the disease stage, the number of tumors was as follows; the 16 (30.76%) tumors were in Clark level I, 10 (19.23%) in Clark level II, 15 (28.84%) in Clark level III, and 11 (21.15%) in Clark level IV and V spontaneously. Among them, five lymph node metastasis, four radial growth phase melanomas (RGPMs), three vertical growth phase melanomas (VGPMs), and 7 Melanoma Metastasis (MelMets) were recorded. The main clinicopathologic data were listed in Table 1.
| Parameter | |||||
| Clark level | I (n=16) | II (n=10) | III (n=15) | IV, V (n=11) | Total (52) |
| Gender (F and M) | F (n=7) | F (n=4) | F (n=5) | F (n=8) | F (n=24) |
| M (n=9) | M (n=6) | M (n=10) | M (n=3) | M (n=28) | |
| Age | 16 (26-79) | 10 (38-68) | 15 (33-89) | 11 (30-70) | 52 (26-89) |
| Location | Face (n=3) | Face (n=3) | Face (n=4) | Face (n=3) | Face (n=13) |
| Hand (n=3) | Hand (n=2) | Palm (n=1) | hand (n=2) | Hand (n=8) | |
| Foot (n=8) | Foot (n=2) | Foot (n=12) | Foot (n=8) | Foot (n=30) | |
| Neck (n=0) | Neck (n=0) | Neck (n=0) | Neck (n=1) | Neck (n=1) | |
| Ulceration | 3 | 2 | 11 | 9 | 25 |
Autophagy markersin different Clark levelsof malignant melanoma
Two crucial genes in autophagosome preparation (ATG5 and BECN1) were checked to analyze the autophagy activity during tumor progression.
Comparison of the ATG5 gene expression level (fold change = 0.8) in melanoma tumors relative to tumor margins (fold change =1) was decreased but not statistically significant (p=0.4). However, the expression pattern differed during tumor progression. The ATG5 expression level was increased in the early stage and then decreased as the stage progressed (p<0.05) (Figure. 1B).
Figure 1. Autophagy Gene Expression in Melanoma Tumors.The gene expression levels of ATG5, BECN1, and GPDH were evaluated in A) melanoma tumors, B) in the early, the intermediate and the advanced stages. The results were represented as fold change and expressed as means ± SD in comparison to tumor margin. The T-test showed significantly difference for *p< 0.05, ** p < 0.01, ***p<0.001.Early stage was Clark level I, intermediate was Clark level II and III and advanced stage was Clark level IV and V.
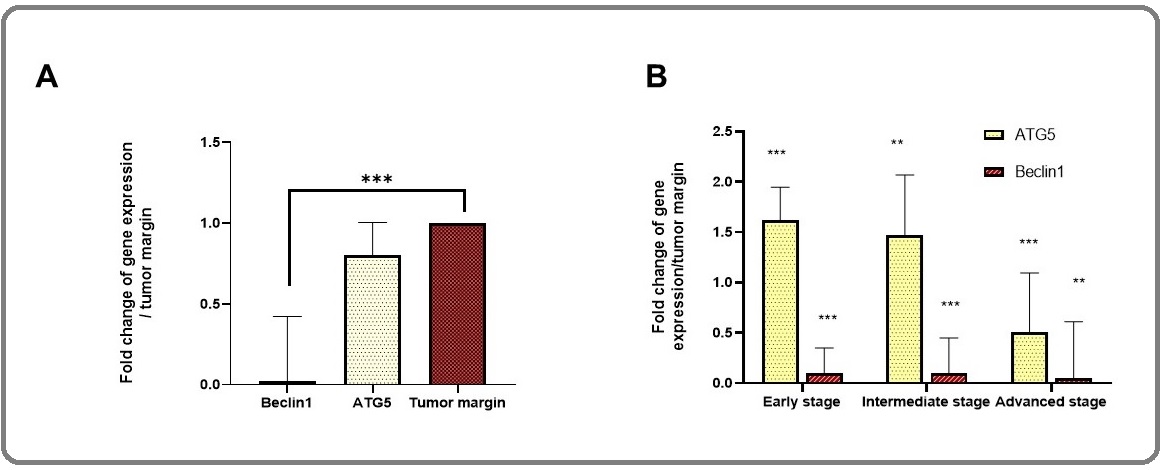
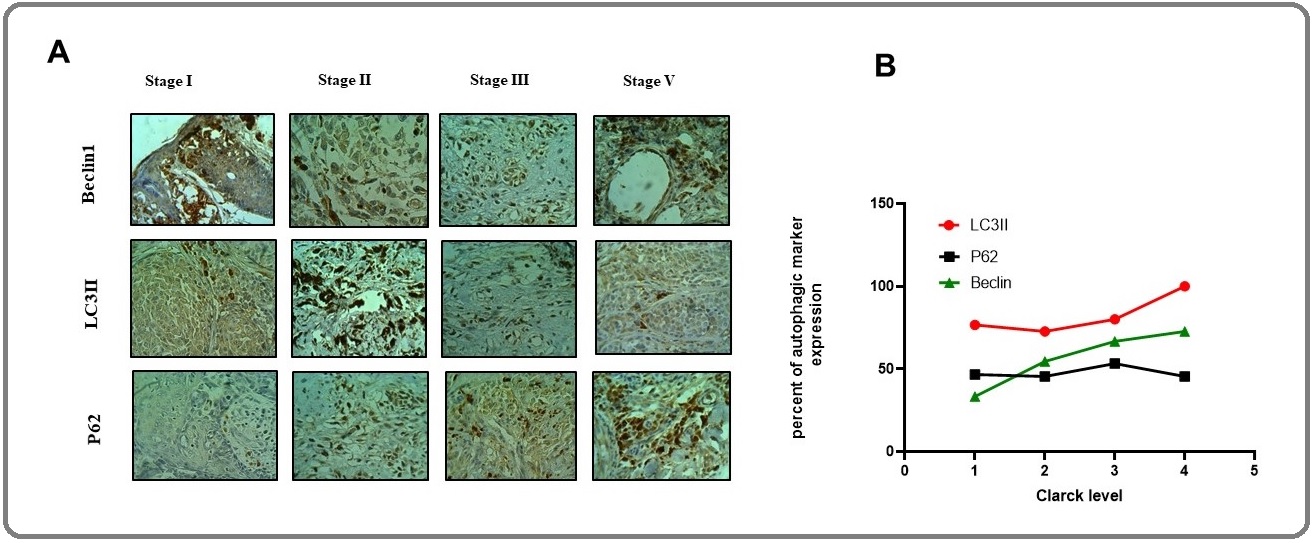
In contrast to ATG5, the expression of BECN1 was significantly reduced in melanoma tumors (fold change = 0.02) compared with tumor margins (fold change =1) with p=0.0001. Indeed, the expression of BECN1 was at the lowest level and not altered during tumor progression (Figure. 1B), where the early stage was considered as Clark level I, the intermediate stage was Clark level II and III and the advanced stage was Clark level IV and V. We subsequently performed immunohistochemistry for BECN1 across a collection of melanoma patients in different stages. Regarding intensity of staining and dot-like staining, analysis of autophagy-related proteins was performed. Figure 2 showed the IHC staining and the expression process of markers based on tumor progression, while Figure 3 illustrated the dot-like staining of markers. In this Figure, the weak, intermediate and strong staining are separated from negative coloring. In talking speech, BECN1 expression was calculated 33/33%, 54/54%, 66/66%, 72/72% in Clark I, II, III, and IV/V, Respectively (Figure 2).
Figure 2. Alterations in Autophagy Protein Levels in Tissue Samples of Melanoma Patients. A) Immunohistochemistry of autophagy marker (BECN1, LC3II, and p62) in different Clark levels of melanoma tumors (400x). B). The quantitative analysis of BECN1, LC3II, and p62 expression in tumor samples compared to tumor margin. Fischer's exact test showed the significance of autophagy marker and Clark level (p<0.05). Weak, intermediate, and strong staining were considered positive.
Totally, BECN1 expression increased with tumor progression (Figure 3). In this regard, the weak, intermediate, and strong staining was considered positive. However, BECN1 expression was relatively highly expressed in melanoma tumors (56.81%) compared with tumor margins (43.18%) with no significant difference (P=0.2).
To test whether ATG5 and BECN1 gene expression during tumor progression results in autophagy induction, we analyzed levels of LC3II and p62 as important protein markers to detect the extent of autophagy. The results showed that the expression of LC3II levels increased as the tumor progressed (Figure 2). In talking speech, the mean percentage of expression was 86/66%, 72/72%, 80%, and 100% in Clark level I, II, III, and IV/V, respectively. We observed markedly higher LC3II levels (mean expression 84.85%) in melanomas compared to tumor margin (mean term 15.15%) (P= 0.001), suggesting increased autophagy in melanoma cells (Figure 2 and 3).
Figure 3. The Level of BECN1, LC3II and, p62 Expression were Evaluated Based on Intensity Score (0: negative, 1: weak, 2: Intermediate, 3: strong). All recordings were calculated in percent compare to tumor margins.
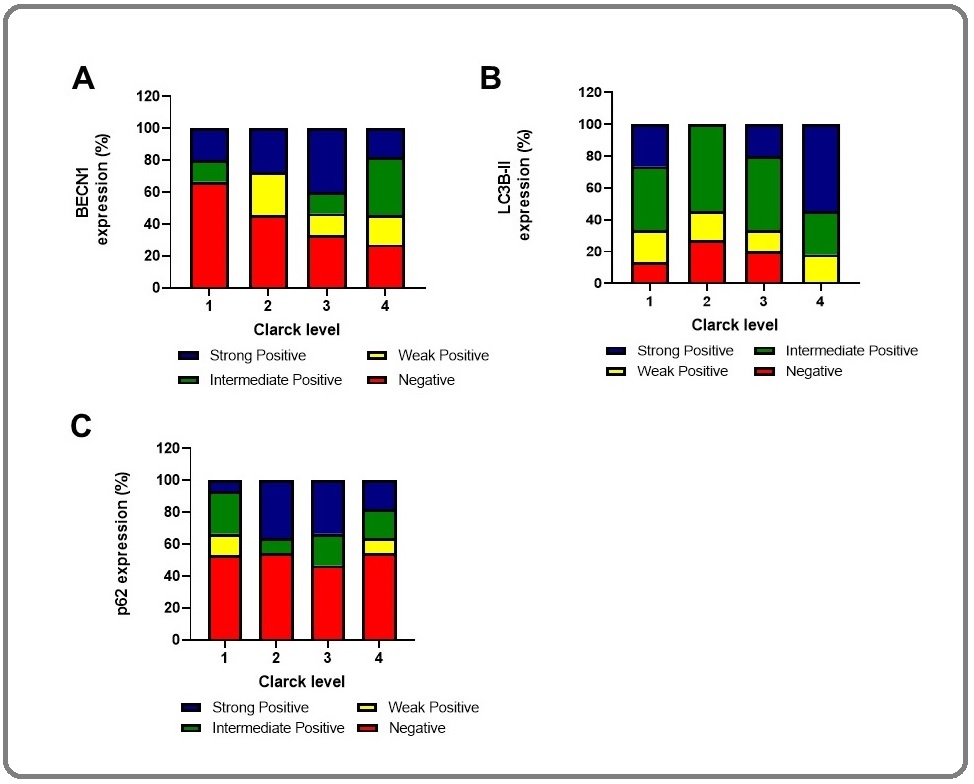
In addition, we analyzed the levels of p62, a marker protein that specifically degraded through autophagy. We observed the mean percentage of p62 expression were 46/66%, 45/45%, 53/33%, and 45/45% in Clark level I, II, III, and IV/V, respectively, which means p62 expression declined as the tumor progressed (Figure 2A, 2B). Indeed, p62 level (mean expression 47.72%) decreased in melanomas, but it was not significantly different in comparison with tumor margin (mean expression 52.27%) (P= 0.1). Altogether, autophagy markers expression in different Clark levels confirmed LC3II and BECN1 protein expression were significantly higher in the late stage of melanoma, whereas the p62 expression decreased when the tumor progressed (Figure 2 and 3). Patients with high BECN1 and LC3 contained a significantly low level of p62 compared with tumor margin indicating autophagy induction in melanoma patients. In other words, BECN1 and LC3II levels correlated inversely with p62 levels in these patients (p<0.05).
Correlation of LC3II and patho-clinical features
The correlation of LC3II dot-like immunohistochemical staining (low and high expression) with patho-clinical features showed that the LC3II didn’t significantly correspond with the location (p=0.373), the thickness of the tumors (p=0.759), the mitotic rate (p=0.795), the lymph node metastasis (p=0.325), and the distant metastasis factors (p=0.331) (Table 2).
| Parameter | LC3II dot-like low (%) | LC3II dot-like high (%) | Total (%) | p-value | |
| Location | Face | 1 (7.7) | 12 (92.3) | 13 (100.0) | 0.373 |
| Hand | 3 (37.5) | 5 (62.5) | 8 (100.0) | ||
| Foot | 8 (26.7) | 22 (73.3) | 30 (100.0) | ||
| Neck | 0 (0.0) | 1 (100.0) | 1 (100.0) | ||
| Ulceration | Absent | 23 (85.18) | 4 (14.81) | 27 (100.0) | 0.032 |
| Present | 4 (16.0) | 21 (84.0) | 25 (100.0) | ||
| Thickness | pTX | 2 (28.6) | 5 (71.4) | 7 (100.0) | 0.759 |
| pT0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| pT1 | 3 (27.3) | 8 (72.7) | 11 (100.0) | ||
| pT2 | 2 (20.0) | 8 (80.0) | 10 (100.0) | ||
| pT3 | 5 (27.8) | 13 (72.2) | 18 (100.0) | ||
| pT4 | 0 (0.0) | 6 (100.0) | 6 (100.0) | ||
| Mitotic rate | none | 4 (26.7) | 11 (73.3) | 15 (100.0) | 0.795 |
| <1 | 3 (21.4) | 11 (21.4) | 14 (100.0) | ||
| 1-5 | 4 (26.7) | 11 (26.7) | 15 (100.0) | ||
| 5-15 | 0 (0.0) | 5 (100.0) | 5 (100.0) | ||
| >15 | 1 (33.3) | 2 (66.7) | 3 (100.0) | ||
| Lymph node metastasis | Absent | 10 (21.3) | 37 (78.7) | 47 (100.0) | 0.325 |
| Present | 2 (40.0) | 3 (60.0) | 5 (100.0) | ||
| Distant metastasis | Absent | 9 (20.0) | 36 (80.0) | 45 (100.0) | 0.331 |
| Present | 3 (42.9) | 4 (57.1) | 7 (100.0) | ||
| Clark level | I | 4 (25.0) | 12 (75.0) | 16 (100.0) | 0.048 |
| II | 3 (30.0) | 7 (70.0) | 10 (100.0) | ||
| III | 4 (26.7) | 11 (73.3) | 15 (100.0) | ||
| IV, V | 1 (9.09) | 10 (90.90) | 11 (100.0) |
However, melanoma anatomic Clark level and ulceration positively match up with LC3II protein expression with p=0.048 and p=0.032, respectively.
Correlation of BECN1 and patho-clinical features
The association of BECN1 dot-like immunohistochemical staining with patho-clinical features showed that the BECN1 didn’t significantly correspond with the location (p=0.135), the thickness of the tumors (p=0.1), the distant metastasis factors (p=0.117). However, BECN1 protein expression positively correlate with the mitotic rate (p=0.005), the lymph node metastasis (p=0.049) melanoma ulceration (p=0.016) and anatomic Clark level (p=0.024) (Table 3).
| Parameter | LC3II dot-like low (%) | LC3II dot-like high (%) | Total (%) | p-value | |
| Location | Face | 6 (46.2) | 7 (53.8) | 13 (100.0) | 0.135 |
| Hand | 7 (87.5) | 1 (12.5) | 8 (100.0) | ||
| Foot | 16 (53.3) | 14 (46.7) | 30 (100.0) | ||
| Neck | 0 (0.0) | 1 (100.0) | 1 (100.0) | ||
| Ulceration | Absent | 25 (92.59) | 2 (7.41) | 27 (100.0) | 0.016 |
| Present | 1 (4.0) | 24 (96.0) | 25 (100.0) | ||
| Thickness | pTX | 4 (57.1) | 3 (42.9) | 7 (100.0) | 0.1 |
| pT0 | 0 (0.0) | 0 (0.0) | 0 (100.0) | ||
| pT1 | 9 (81.8) | 2 (18.2) | 11 (100.0) | ||
| pT2 | 4 (40.0) | 6 (60.0) | 10 (100.0) | ||
| pT3 | 11 (61.1) | 7 (38.9) | 18 (100.0) | ||
| pT4 | 1 (16.7) | 5 (83.3) | 6 (100.0) | ||
| Mitotic rate | none | 7 (46.7) | 8 (53.3) | 15 (100.0) | 0.005 |
| <1 | 13 (92.9) | 1 (7.1) | 14 (100.0) | ||
| 1-5 | 6 (40.0) | 9 (60.0) | 15 (100.0) | ||
| 5-15 | 1 (20.0) | 4 (80.0) | 5 (100.0) | ||
| >15 | 2 (66.7) | 1 (33.3) | 3 (100.0) | ||
| Lymph node metastasis | Absent | 24 (51.1) | 23 (48.9) | 47 (100.0) | 0.049 |
| Present | 5 (100.0) | 0 (0.0) | 5 (100.0) | ||
| Distant metastasis | Absent | 23 (51.1) | 22 (48.9) | 45 (100.0) | 0.117 |
| Present | 6 (85.7) | 1 (14.3) | 7 (100.0) | ||
| Clark level | I | 11 (68.8) | 5 (31.3) | 16 (100.0) | 0.024 |
| II | 7 (70.0) | 3 (30.0) | 10 (100.0) | ||
| III | 7 (46.7) | 8 (53.3) | 15 (100.0) | ||
| Advanced stage | 4 (36.36) | 7 (63.63) | 11 (100.0) |
Correlation of p62 and patho-clinical features
The relation of p62 dot-like immunohistochemical staining with patho-clinical features showed that the p62 didn’t significantly correspond with the location (p=0.642), the thickness of the tumors (p=0.276), the mitotic rate (p=0.264), the lymph node metastasis (p=1.00), and the distant metastasis factors (p=1.00) (Table 4).
| Parameter | LC3II dot-like low (%) | LC3II dot-like high (%) | Total (%) | p-value | |
| Location | Face | 6 (46.2) | 7 (53.8) | 13 (100.0) | 0.642 |
| Hand | 5 (62.5) | 3 (37.5) | 8 (100.0) | ||
| Foot | 18 (60.0) | 12 (40.0) | 30 (100.0) | ||
| Ear | 0 (0.0) | 1 (100.0) | 1 (100.0) | ||
| Ulceration | Absent | 22 (81.48) | 5 (18.51) | 27 (100.0) | 0.04 |
| Present | 3 (12.0) | 22 (88.0) | 25 (100.0) | ||
| Thickness | pTX | 4 (57.1) | 3 (42.9) | 7 (100.0) | 0.276 |
| pT0 | 0 (0.0) | 0 (0.0) | 0 (100.0) | ||
| pT1 | 8 (72.7) | 3 (27.3) | 11 (100.0) | ||
| pT2 | 5 (50.0) | 5 (50.0) | 10 (100.0) | ||
| pT3 | 11 (61.1) | 7 (38.9) | 18 (100.0) | ||
| pT4 | 1 (16.7) | 5 (83.3) | 6 (100.0) | ||
| Mitotic rate | none | 6 (40.0) | 9 (60.0) | 15 (100.0) | 0.264 |
| <1 | 11 (78.6) | 3 (21.4) | 14 (100.0) | ||
| 1-5 | 8 (53.3) | 7 (46.7) | 15 (100.0) | ||
| 5-15 | 2 (40.0) | 3 (60.0) | 5 (100.0) | ||
| >15 | 2 (66.7) | 1 (33.3) | 3 (100.0) | ||
| Lymph node metastasis | Absent | 26 (55.3) | 21 (44.7) | 47 (100.0) | 1 |
| Present | 3 (60.0) | 2 (40.0) | 5 (100.0) | ||
| Distant metastasis | Absent | 25 (55.6) | 20 (44.4) | 45 (100.0) | 1 |
| Present | 4 (57.1) | 3 (42.9) | 7 (100.0) | ||
| Clark level | I | 9 (56.3) | 7 (43.8) | 16 (100.0) | 0.051 |
| II | 6 (60.0) | 4 (40.0) | 10 (100.0) | ||
| III | 7 (46.7) | 8 (53.3) | 15 (100.0) | ||
| IV,V | 7 (63.63) | 4 (36.36) | 11 (100.0) |
However, melanoma ulceration and anatomic Clark level positively match up with p62 protein expression with p=0.040 and p=0.051, respectively.
Biomarker possibility
The analysis of ATG5 and BECN1 gene as the biomarker with Roc curve (graph pad prism 8.0 software) showed that the ATG5 could be a valuable marker with the area under the curve (AUC) 0.7 (p<0.05) against BECN1 with AUC of 0.5 (Figure 4A, 4B).
Figure 4. Roc Curve Analysis of ATG5 and BECN1 Genes. Area under the ROC curve is 0.7 (p<0.02) and 0.5 (p<0.5) for ATG5 and BECN1, respectively.
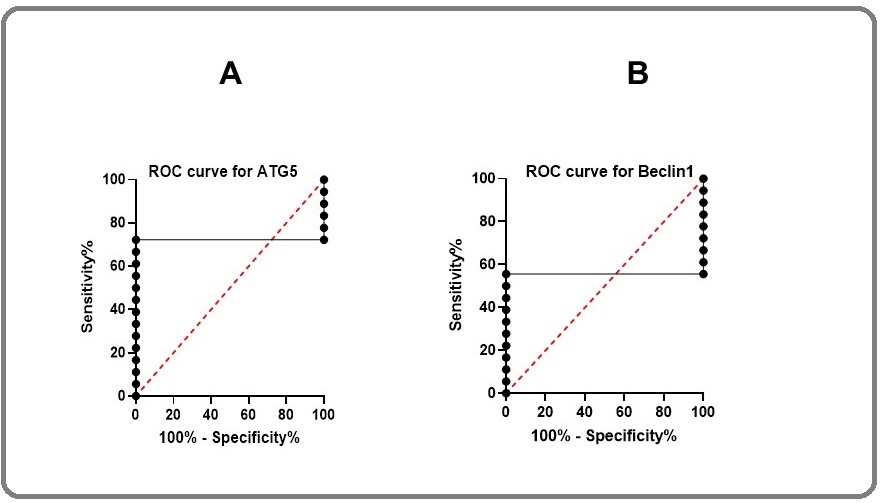
It means that ATG5 would be a reliable marker for melanoma with 70% eventuality.
Discussion
Emerging evidence has shown that there is a correlation between autophagy and tumor progression so that impaired or inhibition of autophagy leads to tumorigenicity. However, different results of autophagy activity during tumorigenesis have been recorded to date [20]. In different cancers, autophagy activity is expected to be low in the early stages while elevated with tumor progression. However, several experiments in melanoma or even other tumors reported controversial results. The reason may be due to misinterpretations of IHC staining or the limitation of IHC for autophagy detection at low or basal levels [21]. For instance, BECN1 acts as a haploinsufficient tumor-suppressor gene and can be either monoallelically deleted or display reduced expression in breast, ovarian, prostatic, and lung cancer [22,23]. In contrast, other reports confirmed that the overexpression of BECN1 correlated with tumorigenesis in colorectal and gastric cancer [24]. The previous study in melanoma tumors showed that BECN1 significantly decreased with tumor progression [25]. In the current experiment, we assessed the BECN1 at both mRNA and protein levels. BECN1 gene expression was reduced in melanoma patients compared to tumor margins (P < 0.0001) and at mRNA level didn’t significantly differ between stages. However, the evaluation of BECN1 level at the protein level showed the decreased level in stage 1 and then went up as the tumor progressed, suggesting an autophagy activity during melanoma progression or autophagy induction may have occurred following chemotherapy which is in agreement with the study of advanced malignant melanoma (MM) that showed overexpression of BECN1 in MM compared to control [26]. Among autophagy-related proteins, LC3II and p62 IHC detection are the most commonly used to study autophagy [27]. LC3II expression has been reported to be either decreased in brain and ovary cancer [28,29] or increased in esophageal and gastrointestinal neoplasms [30]. LC3II expression was associated with poor outcomes in pancreatic cancer and better survival in glioblastoma patients with poor performance scores [28,31]. In cutaneous melanocytic lesions, LC3II significantly decreased with tumor development, and the lowest expression of LC3 II protein was observed in melanoma metastases [20]. While in another experiment in melanoma and breast tissues has confirmed overexpression of LC3 II and the positive correlation with Ki-67 [5]. In the current experimenta high level of LC3 II protein in melanoma tumors was observed compared to tumor margins, suggesting increased autophagy activity during melanoma tumorigenesis.
Overexpression of p62 is also observed in several cancers, including melanoma, and is a marker of poor prognosis. Indeed, increasing levels of p62 expression in the early stage of melanoma followed by subsequent decreasing it, suggesting the novel independent prognostic biomarkers for early-stage melanomas [32]. In this study, p62 monitoring was performed with dot-like staining like the same pattern as BECN1 and LC3II and despite the previous reporting of separate staining of p62 cytoplasm and nucleus [33], we examined both together because no significant difference has been observed and the results confirmed the reduction level of p62 was observed during tumor progression.
Analyzing autophagy in tissues with few markers is very difficult due to the paradoxical function. Therefore, the expression levels of LC3II and p62 markers should be done in parallel with the ATG study [31].There are several reports about the ATG5 gene and protein expression in different cancers, especially melanoma patients. Liu H et al. in 2013 reported ATG5 is often down-regulated in primary melanomas compared to benign nevi. They also checked LC3II, BECN1, and p62 in a few cases. The results have shown that BECN1 was indistinguishable between melanomas and benign nevi, nevertheless a reduced expression of LC3II and increased expression of p62 was observed [34]. In the current experiment, the autophagy markers in different stages of melanoma were determined and revealed that although ATG5 was raised at the early stage, the comparison of ATG5 expression in all melanoma tumors was decreased compared with tumor margin (p<0.05). Autophagy can protect tumor cells exposed to anticancer drugs, indicating the potential for inhibiting autophagy in cancer therapy [35]. It has been shown that elevated autophagy in the early stage is associated with invasiveness and drug resistance [36,37]. Moreover, autophagy was induced in melanoma cells under situations of metastasis [38] and or hypoxia [39]. All these data propose that autophagy may be increased in melanoma cells as an adaptive stress response to chemotherapy and or metabolic stress. In such studies, the quantification of additional ATG proteins should also be considered. So, in addition to ATG5, BECN1 expression at mRNA level was evaluated and revealed that the alteration of expression didn’t observe during tumor progression. However, its expression decreased in all stages compared with tumor margin (p<0.001), suggesting that the expression of ATG proteins is, at least partially, differentially regulated. According to previously published work, a down-regulation of BECN1was revealed in association with melanoma disease progression [20].
In contrast to the previous reports, our experiment in all stages of melanoma showed the autophagy activity during tumor progression. Although in the early stage, a reduction of autophagy may promote tumor growth, by preventing senescence, In later stages, basal autophagy activity resumes or is induced, leading to chemo-resistance. Therefore, the regulation of autophagy in tumorigenesis depends on the stage of the tumors [34].
The prognosis for patients with melanoma depends on variable factors, such as tumor thickness, ulceration, Clark level, and the patient’s age and gender [40]. Among them, ulceration is a predictive factor for the response of adjuvant immune-stimulating therapy [41]. Our experiment showed there is a of BECN1, LC3II and p62 expression levels during tumor progression with ulceration. Although, a previous study reported a direct correlation of melanoma thickness and ulceration in primary melanomas with BECN1non-cytoplasmic expression and inverse with cytoplasmic LC3 II protein expression [20], the importance of autophagy monitoring is demonstrated in melanoma tumor specimens as a valuable prognostic factor.
In conclusion, essential autophagy regulatory proteins, including p62, LC3II, and BECN1, have been proposed as potential prognostic biomarkers providing novel and accurate means to identify tumors at risk of disease progression, facilitating earlier medication intervention, and introducing novel personalized therapeutic approaches to improve clinical outcomes.
Acknowledgments
This study has been funded and supported by Tehran University of Medical Sciences (TUMS), grant no 98- 01-159-41659.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding
This project has been funded and supported by Tehran University of Medical Sciences (TUMS), Tehran, Iran; Grant no: 98-01-159-41659.
Ethical approval
The project was approved by the ethical committee at Tehran University of Medical Sciences, Iran, Code Number: IR.TUMS.VCR.REC.1398.076.
References
- Cutaneous Malignant Melanoma: A Review of Early Diagnosis and Management Naik Piyu Parth. World Journal of Oncology.2021;12(1). CrossRef
- Systematic review of medical treatment in melanoma: current status and future prospects C Garbe, Tk Eigentler, U Keilholz, A Hauschild, Jm Kirkwood. The oncologist.2011;16(1). CrossRef
- WIPI1, BAG1, and PEX3 Autophagy-Related Genes Are Relevant Melanoma Markers D'Arcangelo Daniela, Giampietri Claudia, Muscio Mario, Scatozza Francesca, Facchiano Francesco, Facchiano Antonio. Oxidative Medicine and Cellular Longevity.2018;2018. CrossRef
- Review of diagnostic, prognostic, and predictive biomarkers in melanoma Ankeny Jacob S., Labadie Brian, Luke Jason, Hsueh Eddy, Messina Jane, Zager Jonathan S.. Clinical & Experimental Metastasis.2018;35(5-6). CrossRef
- Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome Lazova Rossitza, Camp Robert L., Klump Vincent, Siddiqui Summar F., Amaravadi Ravi K., Pawelek John M.. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2012;18(2). CrossRef
- An overview of autophagy: morphology, mechanism, and regulation Parzych Katherine R., Klionsky Daniel J.. Antioxidants & Redox Signaling.2014;20(3). CrossRef
- Autophagy: a lysosomal degradation pathway with a central role in health and disease Eskelinen Eeva-Liisa, Saftig Paul. Biochimica Et Biophysica Acta.2009;1793(4). CrossRef
- Atg41/Icy2 regulates autophagosome formation Yao Zhiyuan, Delorme-Axford Elizabeth, Backues Steven K., Klionsky Daniel J.. Autophagy.2015;11(12). CrossRef
- Exploring the Role of Autophagy-Related Gene 5 (ATG5) Yields Important Insights Into Autophagy in Autoimmune/Autoinflammatory Diseases Ye Xin, Zhou Xu-Jie, Zhang Hong. Frontiers in Immunology.2018;9. CrossRef
- The Beclin 1 network regulates autophagy and apoptosis Kang R., Zeh H. J., Lotze M. T., Tang D.. Cell Death and Differentiation.2011;18(4). CrossRef
- Novel Insights into the Cellular Localization and Regulation of the Autophagosomal Proteins LC3A, LC3B and LC3C Baeken Marius W., Weckmann Katja, Diefenthäler Philip, Schulte Jan, Yusifli Kamran, Moosmann Bernd, Behl Christian, Hajieva Parvana. Cells.2020;9(10). CrossRef
- LC3 and Autophagy Tanida Isei, Ueno Takashi, Kominami Eiki. Methods in Molecular Biology (Clifton, N.J.).2008;445. CrossRef
- p62 at the crossroads of autophagy, apoptosis, and cancer Moscat Jorge, Diaz-Meco Maria T.. Cell.2009;137(6). CrossRef
- Physiological significance of selective degradation of p62 by autophagy Komatsu Masaaki, Ichimura Yoshinobu. FEBS letters.2010;584(7). CrossRef
- New insights on the role of autophagy in the pathogenesis and treatment of melanoma Rahmati Marveh, Ebrahim Shiva, Hashemi Saadeh, Motamedi Masoumeh, Moosavi Mohammad Amin. Molecular Biology Reports.2020;47(11). CrossRef
- Statistical analysis of real-time PCR data Yuan Joshua S., Reed Ann, Chen Feng, Stewart C. Neal. BMC bioinformatics.2006;7. CrossRef
- Methods for Evaluating Novel Biomarkers – a New Paradigm Cook Nancy R.. International journal of clinical practice.2010;64(13). CrossRef
- A Method to Reuse Archived H&E Stained Histology Slides for a Multiplex Protein Biomarker Analysis Hinton James P., Dvorak Katerina, Roberts Esteban, French Wendy J., Grubbs Jon C., Cress Anne E., Tiwari Hina A., Nagle Raymond B.. Methods and Protocols.2019;2(4). CrossRef
- Expression analysis of LC3B and p62 indicates intact activated autophagy is associated with an unfavorable prognosis in colon cancer Niklaus Monique, Adams Olivia, Berezowska Sabina, Zlobec Inti, Graber Franziska, Slotta-Huspenina Julia, Nitsche Ulrich, Rosenberg Robert, Tschan Mario P., Langer Rupert. Oncotarget.2017;8(33). CrossRef
- Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions Miracco Clelia, Cevenini Gabriele, Franchi Alessandro, Luzi Pietro, Cosci Elena, Mourmouras Vasileios, Monciatti Irene, Mannucci Susanna, Biagioli Maurizio, Toscano Marzia, Moretti Daniele, Lio Roberto, Massi Daniela. Human Pathology.2010;41(4). CrossRef
- Reliable LC3 and p62 Autophagy Marker Detection in Formalin Fixed Paraffin Embedded Human Tissue by Immunohistochemistry Schläfli Anna, Berezowska Sabina, Adams O., Langer Rupert, Tschan Mario. European Journal of Histochemistry.2015;59. CrossRef
- Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21 Aita V. M., Liang X. H., Murty V. V., Pincus D. L., Yu W., Cayanis E., Kalachikov S., Gilliam T. C., Levine B.. Genomics.1999;59(1). CrossRef
- Decreased expression of Beclin-1 and LC3 in human lung cancer Jiang Zi-Feng, Shao Li-Jie, Wang Wei-Min, Yan Xue-Bo, Liu Rong-Yu. Molecular Biology Reports.2012;39(1). CrossRef
- Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers Ahn Chang Hyeok, Jeong Eun Goo, Lee Jong Woo, Kim Min Sung, Kim Sung Hee, Kim Sung Soo, Yoo Nam Jin, Lee Sug Hyung. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica.2007;115(12). CrossRef
- Beclin-1 and LC3A expression in cutaneous malignant melanomas: a biphasic survival pattern for beclin-1 Sivridis Efthimios, Koukourakis Michael I., Mendrinos Savvas E., Karpouzis Antonios, Fiska Aliki, Kouskoukis Constantinos, Giatromanolaki Alexandra. Melanoma Research.2011;21(3). CrossRef
- Overexpression of autophagy-related beclin-1 in advanced malignant melanoma and its low expression in melanoma-in-situ Hara Yoko, Nakamura Motonobu. European journal of dermatology: EJD.2012;22(1). CrossRef
- Measurement of autophagy in cells and tissues Tanida Isei, Waguri Satoshi. Methods in Molecular Biology (Clifton, N.J.).2010;648. CrossRef
- Monitoring autophagy in glioblastoma with antibody against isoform B of human microtubule-associated protein 1 light chain 3 Aoki Hiroshi, Kondo Yasuko, Aldape Kenneth, Yamamoto Akitsugu, Iwado Eiji, Yokoyama Tomohisa, Hollingsworth E. Faith, Kobayashi Ryuji, Hess Kenneth, Shinojima Naoki, Shingu Takashi, Tamada Yutaka, Zhang Li, Conrad Charles, Bogler Oliver, Mills Gordon, Sawaya Raymond, Kondo Seiji. Autophagy.2008;4(4). CrossRef
- Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer Shen Yang, Li Dan-Dan, Wang Lin-Lin, Deng Rong, Zhu Xiao-Feng. Autophagy.2008;4(8). CrossRef
- Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, et al. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461–8. .
- Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome Fujii Satoshi, Mitsunaga Shuichi, Yamazaki Manabu, Hasebe Takahiro, Ishii Genichiro, Kojima Motohiro, Kinoshita Taira, Ueno Takashi, Esumi Hiroyasu, Ochiai Atsushi. Cancer Science.2008;99(9). CrossRef
- Prognostic impact of p62 expression in cutaneous malignant melanoma Ellis Robert A., Horswell Stuart, Ness Tom, Lumsdon Jonathan, Tooze Sharon A., Kirkham Nigel, Armstrong Jane L., Lovat Penny E.. The Journal of Investigative Dermatology.2014;134(5). CrossRef
- Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu YC, et al . Br J Cancer.2014;111:944-954.
- Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma Liu He, He Zhaoyue, Rütte Thomas, Yousefi Shida, Hunger Robert E., Simon Hans-Uwe. Science Translational Medicine.2013;5(202). CrossRef
- Deconvoluting the context-dependent role for autophagy in cancer White Eileen. Nature Reviews. Cancer.2012;12(6). CrossRef
- Autophagy-deficient mice develop multiple liver tumors Takamura Akito, Komatsu Masaaki, Hara Taichi, Sakamoto Ayako, Kishi Chieko, Waguri Satoshi, Eishi Yoshinobu, Hino Okio, Tanaka Keiji, Mizushima Noboru. Genes & Development.2011;25(8). CrossRef
- Tumour cells resistance in cancer therapy Martínez-Lacaci Isabel, García Morales Pilar, Soto José Luis, Saceda Miguel. Clinical & Translational Oncology: Official Publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico.2007;9(1). CrossRef
- Autophagy Is a Protective Mechanism for Human Melanoma Cells under Acidic Stress Marino Maria Lucia, Pellegrini Paola, Di Lernia Giuseppe, Djavaheri-Mergny Mojgan, Brnjic Slavica, Zhang Xiaonan, Hägg Maria, Linder Stig, Fais Stefano, Codogno Patrice, De Milito Angelo. The Journal of Biological Chemistry.2012;287(36). CrossRef
- Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis Degenhardt Kurt, Mathew Robin, Beaudoin Brian, Bray Kevin, Anderson Diana, Chen Guanghua, Mukherjee Chandreyee, Shi Yufang, Gélinas Céline, Fan Yongjun, Nelson Deirdre A., Jin Shengkan, White Eileen. Cancer Cell.2006;10(1). CrossRef
- Tumor mitotic rate is a more powerful prognostic indicator than ulceration in patients with primary cutaneous melanoma: an analysis of 3661 patients from a single center Azzola Manuela F., Shaw Helen M., Thompson John F., Soong Seng-Jaw, Scolyer Richard A., Watson Geoffrey F., Colman Marjorie H., Zhang Yuting. Cancer.2003;97(6). CrossRef
- Ulcerated Melanoma: Aspects and Prognostic Impact. Cutan Melanoma Etiol Ther Bønnelykke-Behrndtz ML, Steiniche T. Codon Publications.2017;:67-75.
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details