Gene Regulation by p53 in Human Cancer System
Download
Abstract
TP53 proto-oncogene constitutes tumor induction in more than 50% of human cancers as it is mutated frequently in a wide range of cell lines. The transcription of TP53 is postulated to be autoregulated via either binding with TBP and CBF or via direct interaction of p53 protein with TP53 promoter, though further investigation is needed to acknowledge it. Alteration in pathways, regulated through wild type, by mutant p53 (Mutp53) give rise to immortality through interaction with other transcription factors or inducing receptor tyrosine kinases and other signal components. The missense mutation is more frequent constituting more than 60% among all mainly because of the high rate of G>A or C>T transitions in TP53, giving rise to mutation hotspots in R248, R273, etc. In addition to the loss of function, mutations in the TP53 gene also confers oncogenic functions that are not found in wild type p53, referred to as Gain of Function (GOF). GOF mutp53 has been found to promote metastasis, cell proliferation, cell stemness, metabolic reprogramming as well as chemoresistance. Mutp53 also inhibits the wild type effect that is referred to as the Dominant negative effect (DNE). Understanding the mechanisms behind GOF activities, how they promote chemoresistance, and targeting mutp53 will help in improving the treatment of many human cancers with TP53 mutations.
Introduction
In the early 1970s, cancer research was mostly concerned with the cancer-causing viruses that were evidenced to become oncogene. In 1979 the p53 was first discovered as a protein associated with SV40 large T antigen [1]. In 1989, it was first observed that the mutations in TP53 lead to colorectal cancer [2]. It took almost 10 years to realize the wild type p53 as a tumor suppressor protein [2]. p53 acts as a hub node in regulating the normal cell life like DNA damage control, signal transduction, metabolism, cell cycle checkpoints as well as in apoptosis [3]. Wild type p53 binds DNA in a sequence-specific manner while mutant p53 fails to bind to the consensus sequence of wild type target [4,5]. Wide spectrums of genes associated with a range of typical functions performed in cells are somewhat directly or indirectly regulated by the p53 functional domain [6,7]. The genetic variations in TP53, located in chromosome 17p13.1 contribute to human cancers within which the major contributions are of somatic mutations. Germline mutations also to some extent confer on mutant p53 network that affects inheritable mutability through TP53 mutations accumulation. Mutations of p53 in specific regions give rise to cancers in different cell types and not only so, but it also regulates the typical phenotypic expressions of cancer cells to a large extent [8-10]. These conventional, as well as altered networks for both wild type and mutant p53 respectively, make it an interesting gene to study in the realm of molecular oncology.
p53 Domain Structure
The Full-length p53 (FLp53) consists of a total of 393 amino acid residues with distinct functional domains. Functional p53 is a dimer of dimers that are oligomerized to be p53 tetramer through the hydrophobic interactions between Leucine 344 and 348 in the oligomerization domain [11,12] Structurally p53 has five distinct domains, as __N-terminal transactivation domain (TAD), proline-rich domain (PRD), DNA-binding domain (DBD), oligomerization domain (OD), and carboxyl-terminal regulatory domain (CTD) (Figure 1).
Figure 1. Domains of Full-length Protein p53 with Specific Residue Range.

The N-terminus TAD of FLp53 is subdivided into two subdomains as TAD I (1-40 residues) and TAD II (41-67 residues) that can independently activate the transcription of target genes like p21, PUMA. NOXA [13,14]. PRD (68-98 residues) links TAD and DBD with 12 proline residues [14]. DBD (94-292 residues) recognizes and binds specifically targeted DNA consensus sites known as ‘response element’ (RE) [15]. The role of the region sequenced from 293 – 326 is not known [12]. OD (326-353 residues) forms dimer through interaction between helices to form a tetramer [11]. It also contains a Nuclear Export Signal (NES) that is masked by the tetramerization of p53. DNA binding is achieved by interactions between the DBD and OD [16,17]. CTD (353-393 residues) controls the structure and function of the entire protein and is also important in recognizing and binding to damaged DNA by a non-specific interaction. The natively unfolded TADs also interact with various kinds of other proteins like transcription factor II A, II D and II H (TFIIA, TFIID, TFIIH), TATA-box binding protein (TBP), mouse double minute 2 homologs (MDM2) [13,18].In absence of DNA, p53 forms a more loosely arranged cross-like structure though in tight binding with DNA, p53 becomes more rigid and compact [19].
p53 Transcription Auto Regulation and Transcriptional Activation of Several Genes by Wild Type p53
Wild type p53 role is crucial in cell cycle checkpoints in response to DNA damage or in critical cellular stress to prevent tumorigenesis. Upon induction to damage, the TP53 gene transcription is enhanced to form p53 protein that accumulates to interact with Damage Response Factors (DRF), or it targets transcription of p53 target genes through p53 Response Elements (REs) interaction with p53 DNA Binding Domain (DBD) [20,21]. Transcription of wtp53 (Wild type p53) is said to be autoregulated by a feedback mechanism [22]. Two proposed models are conflicting with each other by which p53 can auto-regulate its transcription. One is p53 indirectly interact with TP53 promoter via TATA-box binding protein (TBP) [23,24] or CAAT-box binding factor (CBF) [25]. Wild type p53 interacts with one or more of these proteins to regulate its promoter in a cell type-specific manner [26] (Figure 2B). In another model, it is proposed that p53 directly interacts with TP53 promoter elements and enhances TP53 transcription [22,27] (Figure 2A) .
Figure 2. p53 Transcription is Postulated to be Autoregulated by Either Direct Interaction of p53 Proteins with TP53Promoter (A) or Interactions with TBP, CBF (B), though Further Investigation is Needed. p53 then activates the p53 target gene response element by recruiting Histone acetyltransferase (HAT), Chromatin remodeling complex (CRC), and turn the transcription of target gene on (D). p53 also interacts with Damage response factors (DRF) in DNA damage and helps to choose the repair mechanism (C). p53 also interacts in the pathway of apoptosis which is facilitated by many reasons one of which is a failure in repair phenomenon (E). .
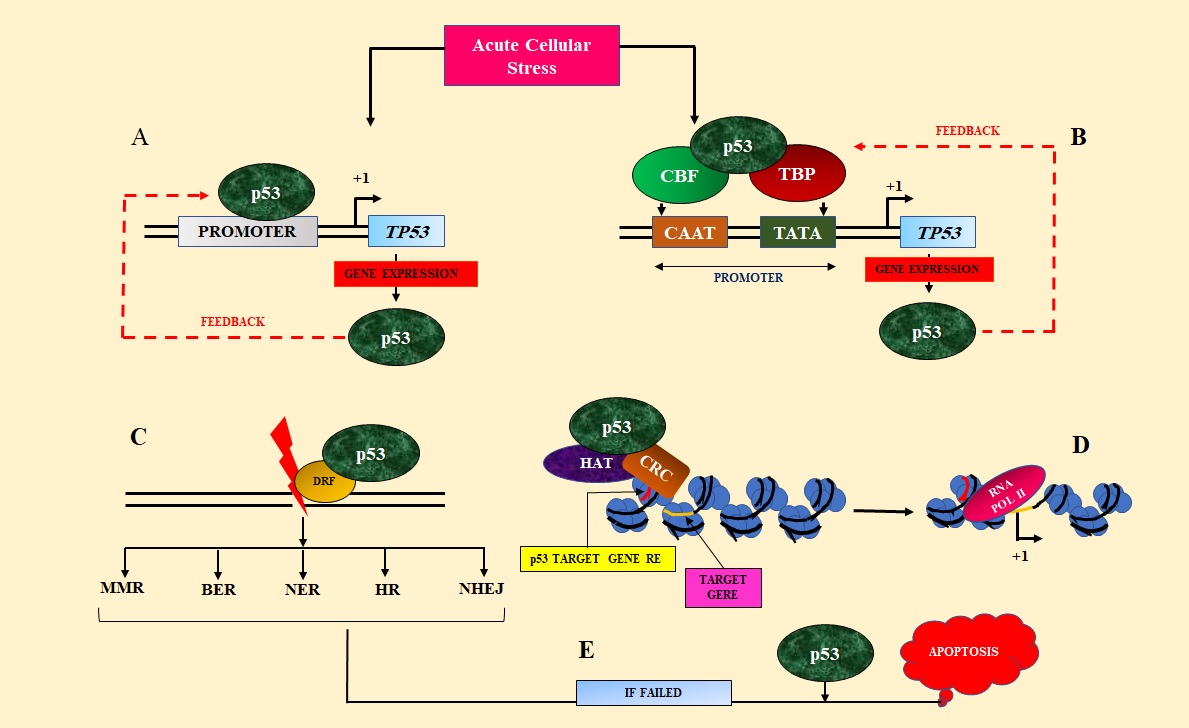
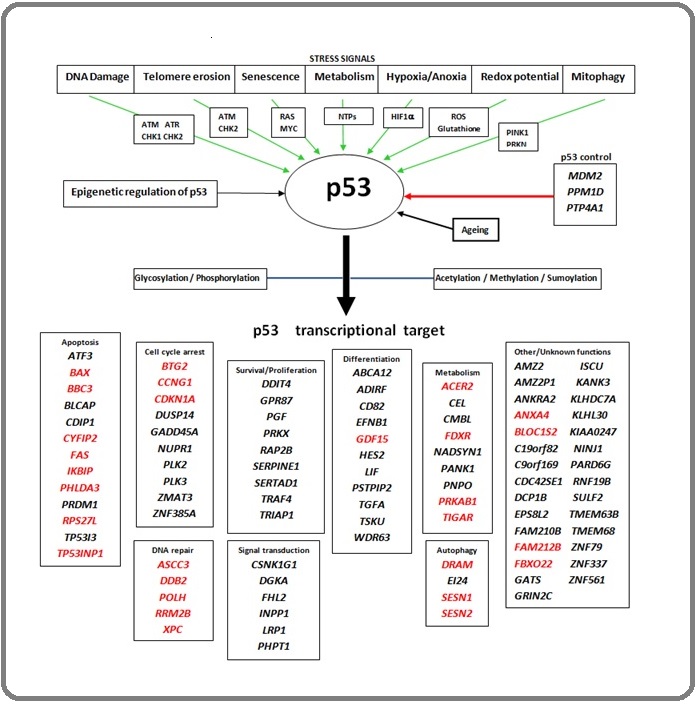
In response to cellular stress signals such as DNA damage, telomere erosion, hypoxia/anoxia, senescence, or mitophagy, wtp53 also activates p53 target gene expression by binding with p53 DNA binding domain preferentially with p53 Response Elements and accumulates because of decreased level of MDM2 (Figure 3).
Figure 3. p53 Regulation Network; Upon Induction through Various Stress Signals Via Different Activating Pathways p53 is Activated (demonstrated via green arrows). It is also negatively regulated via inhibiting control of different gene products (shown via red arrow in p53 controls) and p53 is epigenetically regulated. The nascent p53 becomes functional through post-translational modifications (demonstrated by the blue arrow) to activate p53 transcription target genes that are essential for various important pathways like DNA repair, cell cycle arrest, etc. The genes that are highlighted with red are the early transcriptional target of p53 [24,26] .
Transcription regulatory elements such as transcription factors (TFs), Histone acetyltransferase (HAT) such as p300 are also recruited by wtp53 and acetylates (+Ac) p53 as well as chromatin [20]. This promotes additional steps such as SWI/SNF Chromatin Remodeling Complex (CRC) [28] recruitment that delocalizes Histones in an ATP-dependent manner and RNA Polymerase II can bind to open promoter of p53 target gene and facilitates elongation (Figure 2D). Such a gene is p21 that regulates the expression of cyclin E/ Cyclin-dependent kinase. p53 also interacts with various Damage Response Factors (DRF) such as Replication protein A (RPA), translesion polymerases, etc. Wtp53 also has a potential role to control what kind of repair pathway like Base Excision Repair (BER), Nucleotide Excision Repair (NER), Mismatch Repair (MMR), Homologous Recombination (HR), or Non-homologous End Joining (NHEJ) will induce [21] (Figure 2C). And if all these kinds of repair systems fail to repair the cellular damage, p53 induces apoptosis by interacting with members of BCL-2 family proteins in mitochondria by displacing pro-apoptotic members from pro-apoptotic BCL-2 proteins [21] or by directly activating BAX [29] or BAK to induce Mitochondrial outer membrane permeabilization (MOMP) [21] to release cytochrome c, resulting in programmed cell death (Figure 2E).
Mutation Occurance in p53
Various mutations at specific sites in wild type TP53 converts the tumor suppressor proto-oncogene to oncogene and leads to various kinds of cancer in a wide range of tissue. A wide spectrum of mutations is reported from p53 mutants as Nonsense substitution, Missense substitution, Inframe insertions, Inframe deletions, Frameshift insertions, Frameshift deletions, etc. Data on mutation prevalence in cancer can be easily accessed from the COSMIC database. Analyzing a wide range of unique samples of 48912 reveals that most of the mutations are occurred due to Missense substitution – 30575 (62.3%) and Nonsense substitution - 5187 (10.6%). Also, other kinds of mutations are accumulated in a certain percentage that is observed from the sample study [data are adapted from the COSMIC dataset; https://cancer.sanger.ac.uk/cosmic/gene/analysis? ln=TP53#distribution] (Figure 4A).
Figure 4. Statistical Representation of Data Adopted from COSMIC Database on p53 Mutation Profile. Overall mutation profile (A), transition and transversion (B), transition profile (C) and transversion profile (D) are shown here.
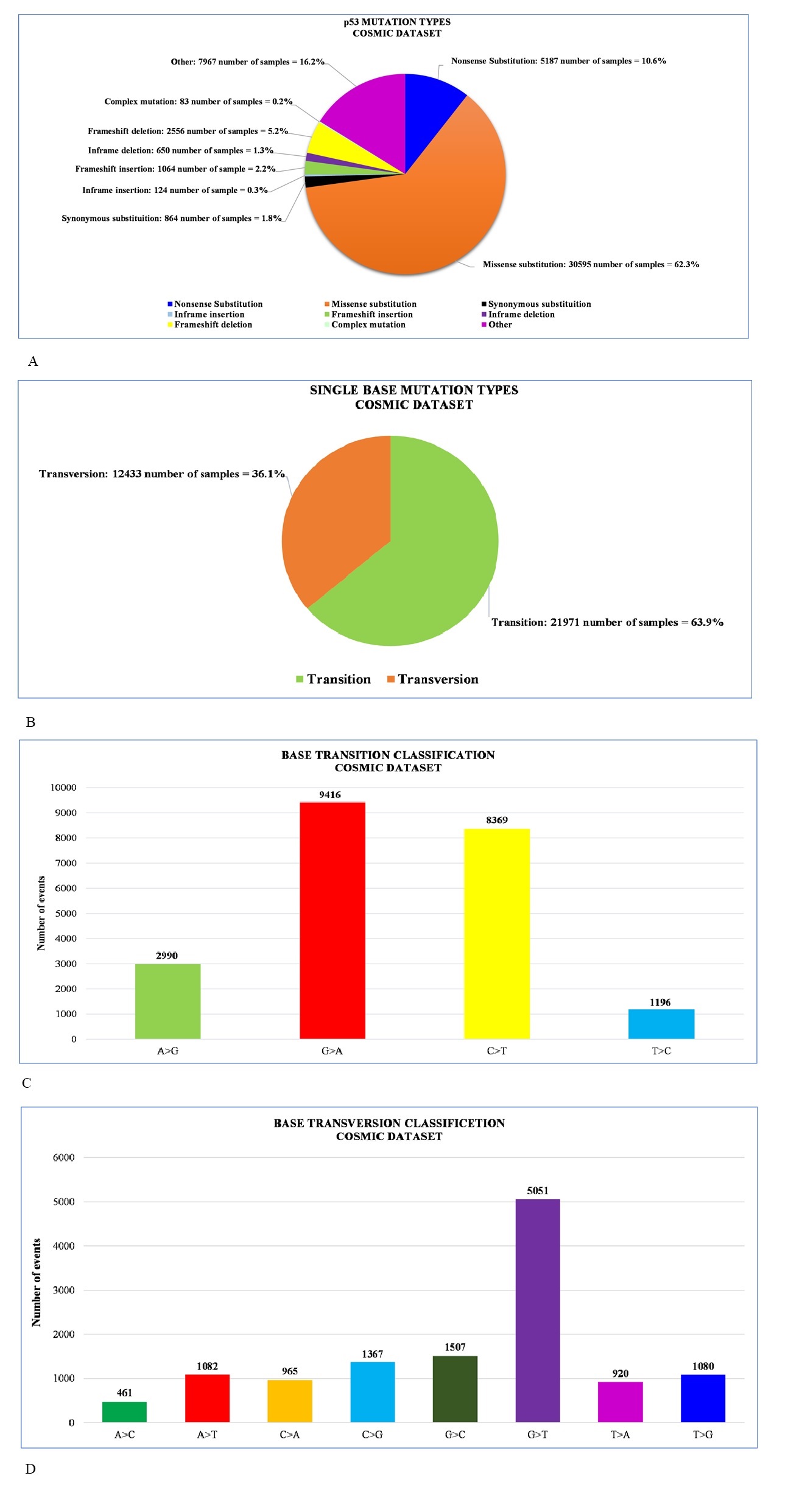
Another study of 32412 unique samples reveals that the rate of transition is much greater (63.9%) than transversions (36.1%) [data are adapted from the COSMIC dataset; https://cancer. sanger.ac.uk/cosmic/gene/analysis?ln=TP53#distribution] (Figure 4B). The G to A and C to T transitions are higher in number constituting about 80.94% of all and the G to T transversions rate is also higher in percentage (40.62%) [data are adapted from COSMIC database; https://cancer. sanger.ac.uk/cosmic/gene/analysis?ln=TP53#distribution] (Figure 4C and 4D).
Mutation hotspots
Majorly the mutations in the DNA binding domain of p53 result in the alteration of structures as well as functions of p53. From the COSMIC dataset mainly ten hotspot regions can be found which include V157F, R175, Y220C, G245, R248, R249, R273, R282 [30] (Figure 5).
Figure 5. Mutation Hotspot in TP53.
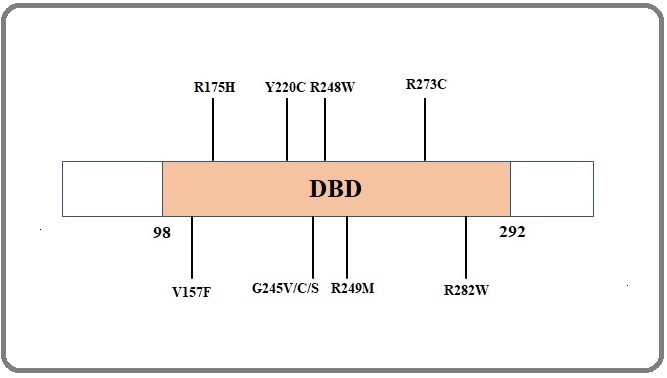
Seven hotspot mutations that are occurred in the DNA binding domain are mostly prevalent (R175H, R248Q, R249S, R248W, R273H, R273C, R282W) over large evolutionary timescales [31]. Some mutagens contribute to the formation of hotspot mutations like solar UV leads to skin cancers (non-melanoma) at hotspot region R248W, R282W, or due to tobacco smoke in regions like G245V, G245C, G245S, R249M [32]. Methylation of CpG residues also leads to the mutation of the TP53 gene and can be explained why the mutation rate at Arginine (R) residue is higher. Four of the total six codons encode for arginine have CpG at the first two positions and U, A, G, or C at the third position and these are the codons that encode arginine due to seven hotspot mutations [30]. The methylated CpG also leads to the higher mutation rate in C to T and G to A mutations are due to antisense of G is methylated C that leads to thymine (T) upon deamination and forming G: C to A: T pairs [30]. Among 22 CpG of DNA-binding domain three hotspot residues (175, 248, 273) represent 60% of CpG mutations, and five other residues (196, 213, 245, 282, 306) represent around 26% of these mutations. Somatic and germline mutations are also prevalent in a huge percentage due to mutations in p53. Somatic TP53 mutations occur in almost every type of cancer in rate from 38%-50% in ovarian, esophageal, colorectal, head and neck, larynx, and lung cancer to about 5% primary leukemia, testicular cancer, cervical cancer, malignant melanoma, etc. [30, 93] and germline mutations are the main cause of Li-Fraumeni Syndrome (LFS) [30,93].
Regulation by Mutant p53 in Broad Spectrum, Leading to Cancer
Faulty transactivation of different p53 target genes, as well as signaling pathways, are mainly mediated due to mutation accumulation in the DNA binding domain (DBD) of p53. In 2017, Pfister et. al. speculated a possible mechanism of mutant p53 transcriptional activity [33]. Despite directly binding with p53 target gene Response Element (RE) mutp53 (mutant p53) interacts with various transcription factors (TFs). Mutant p53 co-opts a set of transcription factors (TFs) that are mainly DNA bound and various co-activators (recruited by either TFs or mutant p53) to initiate gene expression. Though the mechanisms of co-opting individual transcription factors as well as induction and/or activation of cell signaling are broadly unclear and may be dependent on specific p53 mutations. It is also speculated that mutant p53 is involved in the change of chromatin architecture by recruiting known or unknown Chromatin-modifying machinery [41,42]. Mutant p53 also has been reported to induce the receptor tyrosine kinases and other signaling components to promote pro-proliferative signaling [43,44]. The accumulation of hotspot mutations in TP53 facilitates the transcription plasticity and tumor cells increase the capacity of gene expression changes in a particular tumor context (Table 1).
| Domain | Mutations | Cell lines | Associated disease | Ref. |
| DBD | C194D | T47D | Breast Cancer | [34] |
| H193L | CAL27 | Head and Neck Cancer | [34] | |
| M237I | SUM149PT | Breast Cancer | [37] | |
| R110P | SaOS-2 | Osteosarcoma | [35] | |
| R175H | HEK293T | Embryonic kidney | [36] | |
| R175H | SKBr3 | Breast Cancer | [34] | |
| R175H | H1299 | Lung Cancer | [34] | |
| R175H | HCC1395 | Breast Cancer | [37] | |
| R248L | FaDu | Head and neck Cancer | [34] | |
| R248W | MDAH087 | Li-Fraumeni syndrome | [38] | |
| R248W | MIA PaCa-2 | Pancreatic Cancer | [38] | |
| R248W | Colo320 | Colon Cancer | [57] | |
| R249S | BT-549 | Breast Cancer | [37] | |
| R273H | MDA-MB-468 | Breast Cancer | [36] | |
| R273H | U373 | Glioblastoma | [39] | |
| R273H | SNB19 | Glioblastoma | [39] | |
| R273H | HT-29 | Colon Cancer | [40] | |
| R273H | H1299 | Lung Cancer | [34] | |
| R273H | PANC-1 | Pancreatic Cancer | [34] | |
| R273H | MDA-MB-468 | Breast Cancer | [34] | |
| R273H | SW480 and SW620 | Colon Cancer | [57] | |
| R273H | MDA-MB-468 | Breast Cancer | [37] | |
| R280K | MDA-MD-231 | Breast Cancer | [34] |
It is also reported that one p53 mutant behaves differently than other mutants based on their confirmation, binding pattern, cellular localization, or transactivation capacity through p53 gain of function depends on types of cells and tumor context [33].
Dominant Negative Effect of Mutp53 on Wild Type p53
The p53 mutants exert an inhibitory effect on the wild type p53, as a result of which the wild type p53 is prevented from exerting its canonical, normal functions. This is known as the Dominant-negative effect (DNE) (Figure 6) [45].
Figure 6. Dominant Negative Effect of Mutant p53 on Wild-type p53. Mutp53 inhibits the activities of the wild-type p53 to promote tumorigenesis. This phenomenon is referred to as the Dominant-negative-effect of mutp53 on wild-type p53..
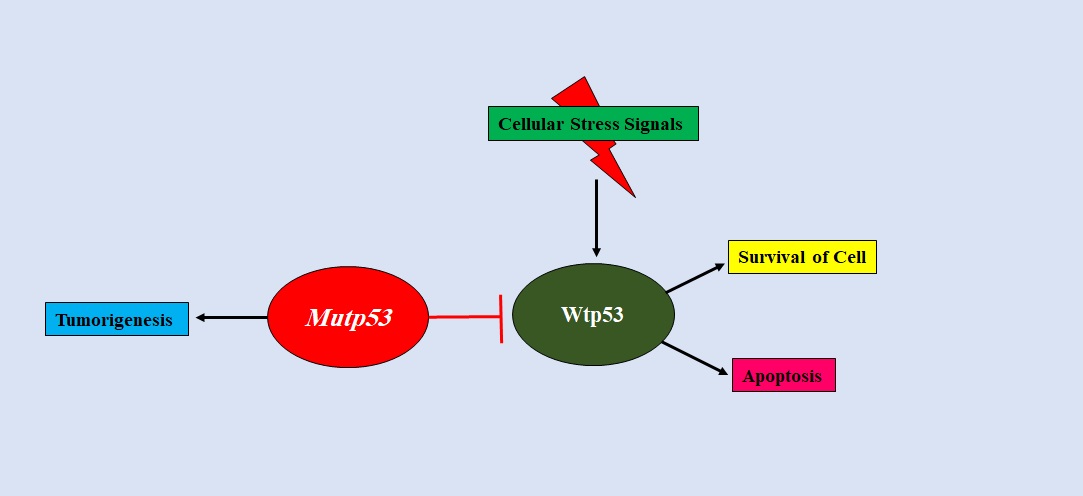
DNE occurs in cancer but its mechanism has not been fully understood. There are several hypotheses out of which one states that mutp53 heterotetramerise with wild type p53 as the tetramerization domain of most p53 DNA binding domain missense mutants are not altered. This heterotetramerisation of mutant p53 with wild-type p53 diminishes the wild-type p53 activity [46]. This is relevant to p53 conformational mutants but not for contact mutants [47]. For contact mutants, it has been proposed that the mutp53 fails to transactivate certain p53 target genes [48]. The wild type/mutant heterotetramer is unable to bind DNA with a high affinity as a wild type p53 homotetramer would [49]. However, there are other possibilities that define the DNE of mutp53. Mutp53 can bind to the transcriptional cofactors required for wild-type p53 activity [50]. The function of wild-type p53 is inhibited by mutp53 via a DNE in a heterozygous condition wherein both wild-type and mutant p53 alleles are present [51].
Gain of Function
Many missense mutp53 are accompanied by new oncogenic functions that are otherwise absent in the wild type p53. These functions help in promoting tumorigenesis. This is referred to as Gain of Function (GOF) [52]. As an instance, when compared with p53 knock-out mice, mutp53 knock-in mice (R172H and R270H) developed more metastatic tumors [53]. Patients with Li-Fraumeni syndrome, carrying germline heterozygous p53 mutations and those with p53 missense mutations develop cancer at an early stage than those patients with p53 deletion mutations [54]. The majority of the p53 mutations that are related to cancer produce proteins that are not truncated and carry only a single amino acid mutation rather than a non-functional truncated protein [55]. GOF mutp53 has been reported to exhibit various functions such as cell proliferation, metastasis, chemoresistance, metabolic reprogramming, cell stemness, etc [8-10].
Mutp53 Assists Cell Proliferation
P53 mutants exhibiting GOF assist cell proliferation, unlike wild-type p53. GOF mutp53 binds to transcription factors such as NF-Y and p300 and promotes the transcription of genes such as cyclin B1, CDK1, CDC25C, thereby stimulating progression through the cell cycle (Figure 7A) [56].
Figure 7. A). Interaction of Mutant p53 with NF-Y and p300. Mutant p53 interacts with HDAC1 independent of DNA damage and suppresses the expression of NF-Y target genes. Upon DNA damage, mutant p53 interacts with p300 and promotes the expression of NF-Y target genes. B). Interaction of mutant p53 with YAP. Mutant p53 interacts with YAP and promotes the expression of proliferative genes. .
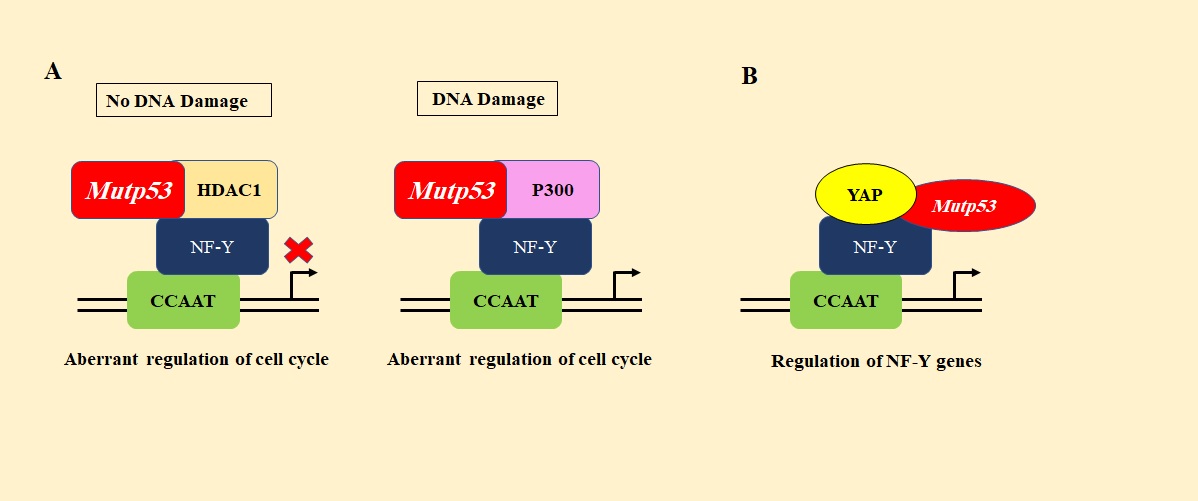
Mutp53 binds to another transcription factor, yes-associated protein (YAP), and stimulates the transcription of cyclin A, CDK1, cyclin B, yet again promoting cell cycle progression and thereby cell proliferation (Figure 7B) [57]. Mutp53 interacts with the promoter of MAP2K3 which, in turn, recruits NF-Y and NFκB to the promoter stimulating MAP2K3 to promote cell cycle progression [58]. In Hepatitis B virus-positive hepatocellular carcinoma, mutant p53-R249S is phosphorylated by CDK4 at Ser249 residue in the G1/S phase. This phosphorylation stimulates its nuclear localization. The phosphorylated p53-R249S interacts with Pin1 and is transported to the nucleus where it binds and stabilizes c-Myc, a nuclear transcriptional factor necessary for proliferation and survival of cancer stem cells, thereby stimulating ribosomal biogenesis and hence promoting hepatocellular carcinoma growth [59]. Furthermore, mutp53 causes the interaction of EGFR and α5β1 with Rab-coupling protein (RCP) and the subsequent translocation of the same to the cell membranes. This stimulates the PI3K/AKT or MAPK cell signaling pathways, ultimately stimulating cell proliferation [9]. R273H mutp53 has also been reported to bind to the promoter region of miR-27a, a negative regulator of EGFR transcript, and suppress its expression, hence activating EGFR/ERK pathways and promoting cell proliferation [82].
Mutp53 Helps in Metastasis
By promoting EMT, mutp53 helps in metastasis. Mutp53 induces ZEB1 expression which is an EMT-related transcription factor, by inhibiting the expression of miR-130b. It directly binds and transrepresses the promoter of miR-130b, thereby stimulating epithelial-mesenchymal transition (EMT) [60]. Mutp53 also induces Twist, another transcription factor of the EMT pathway [61]. Mutp53 forms a complex with p63, a p53 family member and an inhibitor of metastasis, using Smad2/3 (activated by TGFβ) as a molecular bridge between the two proteins (Figure 8A).
Figure 8. A) Inhibition of p63 by Mutant p53 Via Smad2/3. Metastasis is inhibited by p63 but, when it is bound by mutant p53 via the Smad complex, its activity is inhibited, and thereby metastasis is promoted. B) Mutant p53 inhibits de-SUMOylation of Rac1 by SENP1. Upon De-SUMOylation by SENP1, Rac1 is inactivated. This is prevented by mutp53, which, binds to Rac1 and inhibits its de-SUMOylation, thereby promoting migration. .
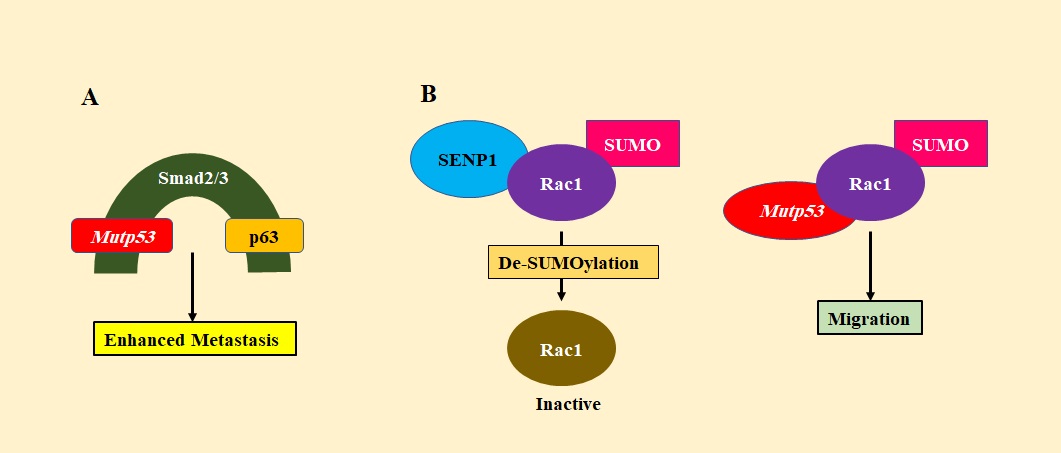
The function of p63 is thereby inhibited and as it cannot upregulate metastatic suppressor genes, EMT is favored [62]. Rac1 is a small GTPase that plays an important role in cell motility and growth. Mutp53 binds to Rac1 and inhibits the interaction of Rac1 with SENP1, thereby inhibiting Rac1 deSUMOylation and activating Rac1 which promotes metastasis (Figure 8B) [63]. Hsp90 alpha isoforms are specially secreted by cancer cells with mutp53. They interact with MMP-2 (matrix metalloproteinase-2), stabilizing it for the degradation of ECM. This helps the cancer cells to metastasize [64]. PDGFRβ (Platelet-derived growth factor receptor β) stimulates various signaling pathways that control cellular movement. p73 binds to NF-Y (transcriptional activators of PDGFRβ) and inhibits it from binding and activating PDGFRβ promoter. Mutp53 disrupts the interaction between p73 and NF-Y and hence activates PDGFRβ, thereby promoting pancreatic cancer metastasis [65].
Mutp53 is Responsible for Metabolic Reprogramming
Mutp53 promotes the uptake of glucose by the cancer cells and the secretion of lactate even under aerobic conditions. This is known as the Warburg effect [66]. Wild-type p53 represses this effect by promoting the expression of genes that are necessary for oxidative phosphorylation [67]. Mutp53 has been found to increase the translocation of GLUT1 to the plasma membrane by activating a GTPase-RhoA and its downstream effector kinase ROCK, thereby enhancing the uptake of glucose and hence glycolysis [68]. Mutp53 also activates the mevalonate pathway. It binds to and activates SREBP (Sterol Regulatory Element-Binding Protein) which is a transcriptional factor that in turn activates HMG-CoA Reductase [69]. Thus, the activation of this pathway also activates RhoA and therefore, YAP or TAZ oncoproteins [70]. Enhancement of anabolic pathways’ activation is important in cancer cells as they help in the formation of macromolecules required for tumor growth. Mutp53 helps in enhancing the activation of anabolic pathways by inhibiting AMPK which is a Ser/Thr kinase. It is activated at high AMP levels, indicating energy stress [71]. Mutp53 binds to the AMPK α subunit and inhibits it. AMPK promotes catabolic pathways and inhibits anabolic pathways [72]. Mutp53 increases the level of reactive oxygen species (ROS) in cancer cells by decreasing glutathione synthesis, resulting in the accumulation of ROS. Wild type p53 activates the TIGAR gene which blocks glycolysis and favors the Pentose phosphate pathway resulting in NADPH production which helps in generating reduced glutathione. By this method, wild-type p53 decreases ROS production [73]. Thus, in the presence of mutp53, there is an increase in oxidative stress. However, cancer cells are protected from such stresses by the same TIGAR gene [74]. Mutp53 also activates the NRF2 gene thereby promoting an oxidative stress survival response that protects the cancer cells from ROS. At the same time, mutp53 represses other targets of NRF2 such as heme oxygenase1 (HMOX1) which has cytotoxic effects on cancer cells but protects the normal cells [75].
Cell Stemness is Promoted by Mutp53
A study proposed that cancer stemness is promoted by mutp53 in glioblastoma and breast cancer cells by activating the PI3K/AKT2 mediated growth factor receptor cycling. AKT2 in turn phosphorylates WIP (WASP-interacting protein) which stabilizes YAP/TAZ thereby supporting CSC survival [76]. Further, mutp53 interacts with SREBP and activates the mevalonate pathway. This pathway produces geranylgeranyl pyrophosphate which activates a small GTPase, Rho which in turn activates YAP/TAZ thereby promoting self-renewal of breast cancer cells [70]. In hematopoietic stem cells, mutp53 promoted an increased ability to self-renew by upregulating FoxH1, which is a regulator of stem cell factor receptor c-kit and SCA-1 (Stem Cell Antigen 1) [77].
Mutp53- The Culprit Behind Failed Cancer Therapies
GOF mutp53 promotes resistance to anticancer drugs and therapies and plays a major role in cancer-related death [78]. Among patients with breast cancer, it was observed that those with mutp53 had a lower chance of survival than those with wild-type p53 tumors [79]. In carcinoma cells, it was found that mutp53 provides resistance to various chemotherapeutic drugs like cisplatin, doxorubicin, and 5-fluorouracil by inhibiting apoptosis via procaspase-3 repression [80]. In breast cancer with mutant p53, it was shown that through the inhibition of miR-30c and REV1, resistance to adriamycin was achieved indicating that mutp53 favors the DNA damage repair (DDR) pathway. Chemotherapeutic drugs induce DNA damage in cancer cells and therefore promoting the DDR pathway provides resistance against these drugs [81]. Studies have shown that mutp53 increases the resistance of glioblastoma to temozolomide by increasing MGMT expression (O6-methyl guanine DNA-methyltransferase), which is an enzyme involved in the repair of DNA damaged by the drug temozolomide [82]. It has been reported recently that mutp53 affects EFNB2, a receptor tyrosine kinase involved in the regulation of migration, invasion, and tumor resistance [83]. When colorectal carcinoma cells were treated with 5-FU, mutp53 increased EFNB2 expression. EFNB2, in turn, upregulates ABCG2 (ATP-binding cassette sub-family G2), a multidrug resistance efflux transporter, via the activation of the JNK pathway [84]. When Ca2+ is transferred from the endoplasmic reticulum to the mitochondria, this causes pro-apoptotic responses. If this process is interfered with, the chances of cell survival increases. Under stress conditions, wild-type p53 increases the transfer of Ca2+ from the endoplasmic reticulum to mitochondria, promoting apoptosis [85]. In the presence of mutp53, Ca2+ is not transferred from the endoplasmic reticulum of cancer cells to the mitochondria, promoting chemoresistance to stressful treatments [86]. In colorectal cancer, it was shown that mutp53-R273H provides resistance against 5-FU by downregulating the proapoptotic protein PUMA, which is an activator of BAX, a proapoptotic protein. Mutp53 was unable to bind to the PUMA promoter, thereby inhibiting its transcription. Thus, mutp53 decreases the apoptotic activity of BAX [87]. Mutp53 provides chemoresistance by inducing the expression of Cytochrome P450 (CYP450) family members such as CYP3A4. This enzyme metabolizes several chemotherapeutic drugs thus providing cancer cells resistance against them [88]. In conclusions, P53 protein is known as the ‘Guardian of the genome’ as it regulates the expression of several genes involved in DNA repair, cell growth, apoptosis, and senescence. It is therefore a tumor suppressor gene [89]. However, when the TP53 gene is mutated, it promotes tumorigenesis via its GOF activities [52]. It is mutated in more than 50% of human cancers and is, therefore, an important target for cancer therapy [90]. Targeting mutp53 comes with a lot of challenges, one of which is reactivating wtp53 as it is a tumor suppressor gene. Unfortunately, this is a difficult task as it is easier to inhibit a protein than to activate it [91]. Thus, to treat cancers with TP53 mutations, new strategies are necessary [92]. In terms of their biological effects, TP53 mutations are diverse and may take place at different stages of tumor development. This makes it extremely difficult to extract information for clinical purposes [93]. Since p53 plays a major role in cancer biology, strategies are required that activate apoptosis mediated by p53 and also suppress the dominant-negative effect of mutp53 on wtp53 [94]. The role of MDM2 is to inhibit p53 as well as cause the proteasomal breakdown of the protein. Thus, as a new therapeutic approach, an MDM2 antagonist can be used to activate p53. Nutlin, an inhibitor of p53-MDM2 binding was discovered by Vassilev et al. As a result of this, p53 is stabilized and p53- mediated apoptotic pathway is activated in cancer cells with wtp53 [95]. Additionally, an inhibitor of MDM2, M1-219 has also been shown to be an effective agent in reactivating p53 [96]. Thus, using such agents in cancer therapy and developing various strategies that reactivate p53 is required for treating malignant tumors with mutp53, effectively [92]. Unfortunately, there are several unresolved areas in cancer therapy targeting mutp53. More studies are required in this field which would eventually, in the future provide effective, efficient, and precise cancer treatments targeting mutp53 [68]. Although a lot of progress has been made in the studies of the role of mutp53 in cancer, all the information is still very limited. More research on the same and the mechanism of mutp53’s GOF activities as well as the dominant-negative effect of mutp53 on wtp53 will be the next big achievement in cancer research.
Authors’ contribution
Blueprint and layout, Abstract: SG and MB; SG writes the manuscript part from Introduction to ‘Regulation by mutant p53 in broad spectrum, leading to cancer’; MB writes the manuscript part from ‘Dominant negative effect of mutp53 on wild type p53’ to Conclusions; all the figures of respective areas are created by the authors not adapted directly from anywhere. NKJ corrected and guided everything. All authors have read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
Authors are grateful to Department of Life Science and Biotechnology, Jadavpur University for supporting SG and MB, and Department of Biotechnology, Heritage Institute of Technology for supporting NKJ. Authors are also thankful to Dr. Paltu Kumar Dhal, Assistant Professor, Department of Life Science and Biotechnology, Jadavpur University for informing about COSMIC database.
References
- T antigen is bound to a host protein in SV40-transformed cells Lane D. P., Crawford L. V.. Nature.1979;278(5701). CrossRef
- Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas Baker S. J., Fearon E. R., Nigro J. M., Hamilton S. R., Preisinger A. C., Jessup J. M., vanTuinen P., Ledbetter D. H., Barker D. F., Nakamura Y., White R., Vogelstein B.. Science (New York, N.Y.).1989;244(4901). CrossRef
- Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Bargonetti J., Reynisdóttir I., Friedman P. N., Prives C.. Genes & Development.1992;6(10). CrossRef
- Definition of a consensus binding site for p53 Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B.. Nature Genetics.1992;1(1). CrossRef
- A transcriptionally active DNA-binding site for human p53 protein complexes Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Molecular and Cellular Biology.1992;12(6). CrossRef
- Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity Andrysik Zdenek, Galbraith Matthew D., Guarnieri Anna L., Zaccara Sara, Sullivan Kelly D., Pandey Ahwan, MacBeth Morgan, Inga Alberto, Espinosa Joaquín M.. Genome Research.2017;27(10). CrossRef
- Tied Up in Loops: Positive and Negative Autoregulation of p53 Lu Xin. Cold Spring Harbor Perspectives in Biology.2009. CrossRef
- Mutant p53 Gain-of-Function: Role in Cancer Development, Progression, and Therapeutic Approaches Alvarado-Ortiz Eduardo, Cruz-López Karen Griselda, Becerril-Rico Jared, Sarabia-Sánchez Miguel Angel, Ortiz-Sánchez Elizabeth, García-Carrancá Alejandro. Frontiers in Cell and Developmental Biology.2020;8. CrossRef
- Mutant p53 drives invasion by promoting integrin recycling Muller Patricia A. J., Caswell Patrick T., Doyle Brendan, Iwanicki Marcin P., Tan Ee H., Karim Saadia, Lukashchuk Natalia, Gillespie David A., Ludwig Robert L., Gosselin Pauline, Cromer Anne, Brugge Joan S., Sansom Owen J., Norman Jim C., Vousden Karen H.. Cell.2009;139(7). CrossRef
- Understanding the Intersections between Metabolism and Cancer Biology Vander Heiden Matthew G., DeBerardinis Ralph J.. Cell.2017;168(4). CrossRef
- Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms Jeffrey P. D., Gorina S., Pavletich N. P.. Science (New York, N.Y.).1995;267(5203). CrossRef
- Structural biology of the tumor suppressor p53 Joerger Andreas C., Fersht Alan R.. Annual Review of Biochemistry.2008;77. CrossRef
- Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein Chang J., Kim D. H., Lee S. W., Choi K. Y., Sung Y. C.. The Journal of Biological Chemistry.1995;270(42). CrossRef
- Identification of a novel p53 functional domain that is necessary for efficient growth suppression Walker K. K., Levine A. J.. Proceedings of the National Academy of Sciences of the United States of America.1996;93(26). CrossRef
- Transcriptional control of human p53-regulated genes Riley Todd, Sontag Eduardo, Chen Patricia, Levine Arnold. Nature Reviews. Molecular Cell Biology.2008;9(5). CrossRef
- The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain D'Abramo M., Bešker N., Desideri A., Levine A. J., Melino G., Chillemi G.. Oncogene.2016;35(25). CrossRef
- p53–RNA interactions: New clues in an old mystery Riley Kasandra J.-L., Maher L. James. RNA.2007. CrossRef
- Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53 Di Lello Paola, Jenkins Lisa M. Miller, Jones Tamara N., Nguyen Bao D., Hara Toshiaki, Yamaguchi Hiroshi, Dikeakos Jimmy D., Appella Ettore, Legault Pascale, Omichinski James G.. Molecular Cell.2006;22(6). CrossRef
- Structural Evolution and Dynamics of the p53 Proteins Chillemi Giovanni, Kehrloesser Sebastian, Bernassola Francesca, Desideri Alessandro, Dötsch Volker, Levine Arnold J., Melino Gerry. Cold Spring Harbor Perspectives in Medicine.2017;7(4). CrossRef
- Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., Halazonetis T. D., Berger S. L.. Molecular Cell.2001;8(6). CrossRef
- How the Other Half Lives: What p53 Does When It Is Not Being a Transcription Factor Ho Teresa, Tan Ban Xiong, Lane David. International Journal of Molecular Sciences.2020;21(1). CrossRef
- p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints Wang Shulin, El-Deiry Wafik S.. Cancer Research.2006;66(14). CrossRef
- Wild-type p53 binds to the TATA-binding protein and represses transcription Seto E., Usheva A., Zambetti G. P., Momand J., Horikoshi N., Weinmann R., Levine A. J., Shenk T.. Proceedings of the National Academy of Sciences of the United States of America.1992;89(24). CrossRef
- “Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein.” Truant R, et al . The Journal of biological chemistry.1993;268(4):2284-2287.
- Regulation of the human hsp70 promoter by p53 Agoff S. N., Hou J., Linzer D. I., Wu B.. Science (New York, N.Y.).1993;259(5091). CrossRef
- Wild-type p53 regulates its own transcription in a cell-type specific manner Hudson J. M., Frade R., Bar-Eli M.. DNA and cell biology.1995;14(9). CrossRef
- The tumor suppressor p53 regulates its own transcription Deffie A., Wu H., Reinke V., Lozano G.. Molecular and Cellular Biology.1993;13(6). CrossRef
- SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription Lee Daeyoup, Kim Jin Woo, Seo Taegun, Hwang Sun Gwan, Choi Eui-Ju, Choe Joonho. The Journal of Biological Chemistry.2002;277(25). CrossRef
- A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine bax genes Thornborrow Edward C., Patel Sejal, Mastropietro Anthony E., Schwartzfarb Elissa M., Manfredi James J.. Oncogene.2002;21(7). CrossRef
- Why are there hotspot mutations in the TP53 gene in human cancers? Baugh Evan H., Ke Hua, Levine Arnold J., Bonneau Richard A., Chan Chang S.. Cell Death & Differentiation.2018;25(1). CrossRef
- Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database Petitjean Audrey, Mathe Ewy, Kato Shunsuke, Ishioka Chikashi, Tavtigian Sean V., Hainaut Pierre, Olivier Magali. Human Mutation.2007;28(6). CrossRef
- Somatic TP53 Mutations in the Era of Genome Sequencing Hainaut Pierre, Pfeifer Gerd P.. Cold Spring Harbor Perspectives in Medicine.2016;6(11). CrossRef
- Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53 Pfister Neil T., Prives Carol. Cold Spring Harbor Perspectives in Medicine.2017;7(2). CrossRef
- Mutant p53 cooperates with ETS2 to promote etoposide resistance Do Phi M., Varanasi Lakshman, Fan Songqing, Li Chunyang, Kubacka Iwona, Newman Virginia, Chauhan Krishna, Daniels Silvano Rakeem, Boccetta Maurizio, Garrett Michael R., Li Runzhao, Martinez Luis A.. Genes & Development.2012;26(8). CrossRef
- Physical interaction with human tumor-derived p53 mutants inhibits p63 activities Strano Sabrina, Fontemaggi Giulia, Costanzo Antonio, Rizzo Maria Giulia, Monti Olimpia, Baccarini Alessia, Del Sal Giannino, Levrero Massimo, Sacchi Ada, Oren Moshe, Blandino Giovanni. The Journal of Biological Chemistry.2002;277(21). CrossRef
- Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer Walerych Dawid, Lisek Kamil, Sommaggio Roberta, Piazza Silvano, Ciani Yari, Dalla Emiliano, Rajkowska Katarzyna, Gaweda-Walerych Katarzyna, Ingallina Eleonora, Tonelli Claudia, Morelli Marco J., Amato Angela, Eterno Vincenzo, Zambelli Alberto, Rosato Antonio, Amati Bruno, Wiśniewski Jacek R., Del Sal Giannino. Nature Cell Biology.2016;18(8). CrossRef
- ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters Dell'Orso Stefania, Fontemaggi Giulia, Stambolsky Perry, Goeman Frauke, Voellenkle Christine, Levrero Massimo, Strano Sabrina, Rotter Varda, Oren Moshe, Blandino Giovanni. Omics: A Journal of Integrative Biology.2011;15(5). CrossRef
- A novel PTEN/mutant p53/c-Myc/Bcl-XL axis mediates context-dependent oncogenic effects of PTEN with implications for cancer prognosis and therapy Huang Xiaoping, Zhang Ying, Tang Yaqiong, Butler Napoleon, Kim Jungeun, Guessous Fadila, Schiff David, Mandell James, Abounader Roger. Neoplasia (New York, N.Y.).2013;15(8). CrossRef
- Wild-type p53 controls cell motility and invasion by dual regulation of MET expression Hwang Chang-Il, Matoso Andres, Corney David C., Flesken-Nikitin Andrea, Körner Stefanie, Wang Wei, Boccaccio Carla, Thorgeirsson Snorri S., Comoglio Paolo M., Hermeking Heiko, Nikitin Alexander Yu.. Proceedings of the National Academy of Sciences.2011;108(34). CrossRef
- A common polymorphism acts as an intragenic modifier of mutant p53 behaviour Marin M. C., Jost C. A., Brooks L. A., Irwin M. S., O'Nions J., Tidy J. A., James N., McGregor J. M., Harwood C. A., Yulug I. G., Vousden K. H., Allday M. J., Gusterson B., Ikawa S., Hinds P. W., Crook T., Kaelin W. G.. Nature Genetics.2000;25(1). CrossRef
- Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells Ali Amjad, Wang Zhuo, Fu Junjiang, Ji Lei, Liu Jiang, Li Lei, Wang Hui, Chen Jiwu, Caulin Carlos, Myers Jeffrey N., Zhang Pei, Xiao Jianru, Zhang Bianhong, Li Xiaotao. Nature Communications.2013;4. CrossRef
- Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells Pfister Neil T., Fomin Vitalay, Regunath Kausik, Zhou Jeffrey Y., Zhou Wen, Silwal-Pandit Laxmi, Freed-Pastor William A., Laptenko Oleg, Neo Suat Peng, Bargonetti Jill, Hoque Mainul, Tian Bin, Gunaratne Jayantha, Engebraaten Olav, Manley James L., Børresen-Dale Anne-Lise, Neilsen Paul M., Prives Carol. Genes & Development.2015;29(12). CrossRef
- Cell signaling by receptor tyrosine kinases Lemmon Mark A., Schlessinger Joseph. Cell.2010;141(7). CrossRef
- Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation Marshall C. J.. Cell.1995;80(2). CrossRef
- When mutants gain new powers: news from the mutant p53 field Brosh Ran, Rotter Varda. Nature Reviews. Cancer.2009;9(10). CrossRef
- Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation Milner Jo, Medcalf E. A.. Cell.1991;65(5). CrossRef
- In vitro analysis of the dominant negative effect of p53 mutants Chène P.. Journal of Molecular Biology.1998;281(2). CrossRef
- Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes Willis Amy, Jung Eun Joo, Wakefield Therese, Chen Xinbin. Oncogene.2004;23(13). CrossRef
- UV-dependent alternative splicing uncouples p53 activity and PIG3 gene function through rapid proteolytic degradation Nicholls Chris D., Shields Michael A., Lee Patrick W. K., Robbins Stephen M., Beattie Tara L.. The Journal of Biological Chemistry.2004;279(23). CrossRef
- p53 modulation of TFIIH-associated nucleotide excision repair activity Wang X. W., Yeh H., Schaeffer L., Roy R., Moncollin V., Egly J. M., Wang Z., Freidberg E. C., Evans M. K., Taffe B. G.. Nature Genetics.1995;10(2). CrossRef
- Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis Rivlin Noa, Brosh Ran, Oren Moshe, Rotter Varda. Genes & Cancer.2011;2(4). CrossRef
- Gain of function mutations in p53 Dittmer D., Pati S., Zambetti G., Chu S., Teresky A. K., Moore M., Finlay C., Levine A. J.. Nature Genetics.1993;4(1). CrossRef
- Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome Lang Gene A., Iwakuma Tomoo, Suh Young-Ah, Liu Geng, Rao V. Ashutosh, Parant John M., Valentin-Vega Yasmine A., Terzian Tamara, Caldwell Lisa C., Strong Louise C., El-Naggar Adel K., Lozano Guillermina. Cell.2004;119(6). CrossRef
- Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families Bougeard G., Sesboüé R., Baert-Desurmont S., Vasseur S., Martin C., Tinat J., Brugières L., Chompret A., Paillerets B. Bressac, Stoppa-Lyonnet D., Bonaïti-Pellié C., Frébourg T.. Journal of Medical Genetics.2008;45(8). CrossRef
- p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells Rotter V.. Proceedings of the National Academy of Sciences of the United States of America.1983;80(9). CrossRef
- Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation Di Agostino Silvia, Strano Sabrina, Emiliozzi Velia, Zerbini Valentina, Mottolese Marcella, Sacchi Ada, Blandino Giovanni, Piaggio Giulia. Cancer Cell.2006;10(3). CrossRef
- YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins Di Agostino Silvia, Sorrentino Giovanni, Ingallina Eleonora, Valenti Fabio, Ferraiuolo Maria, Bicciato Silvio, Piazza Silvano, Strano Sabrina, Del Sal Giannino, Blandino Giovanni. EMBO reports.2016;17(2). CrossRef
- Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function Gurtner Aymone, Starace Giuseppe, Norelli Giuseppe, Piaggio Giulia, Sacchi Ada, Bossi Gianluca. The Journal of Biological Chemistry.2010;285(19). CrossRef
- Mutant p53 Gains Its Function via c-Myc Activation upon CDK4 Phosphorylation at Serine 249 and Consequent PIN1 Binding Liao Peng, Zeng Shelya X., Zhou Xiang, Chen Tianjian, Zhou Fen, Cao Bo, Jung Ji Hoon, Del Sal Giannino, Luo Shiwen, Lu Hua. Molecular Cell.2017;68(6). CrossRef
- Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis Dong P., Karaayvaz M., Jia N., Kaneuchi M., Hamada J., Watari H., Sudo S., Ju J., Sakuragi N.. Oncogene.2013;32(27). CrossRef
- Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells Kogan-Sakin I., Tabach Y., Buganim Y., Molchadsky A., Solomon H., Madar S., Kamer I., Stambolsky P., Shelly A., Goldfinger N., Valsesia-Wittmann S., Puisieux A., Zundelevich A., Gal-Yam E. N., Avivi C., Barshack I., Brait M., Sidransky D., Domany E., Rotter V.. Cell Death and Differentiation.2011;18(2). CrossRef
- A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis Adorno Maddalena, Cordenonsi Michelangelo, Montagner Marco, Dupont Sirio, Wong Christine, Hann Byron, Solari Aldo, Bobisse Sara, Rondina Maria Beatrice, Guzzardo Vincenza, Parenti Anna R., Rosato Antonio, Bicciato Silvio, Balmain Allan, Piccolo Stefano. Cell.2009;137(1). CrossRef
- Gain-of-function mutant p53 activates small GTPase Rac1 through SUMOylation to promote tumor progression Yue Xuetian, Zhang Cen, Zhao Yuhan, Liu Juan, Lin Alan W., Tan Victor M., Drake Justin M., Liu Lianxin, Boateng Michael N., Li Jun, Feng Zhaohui, Hu Wenwei. Genes & Development.2017;31(16). CrossRef
- Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness Eustace Brenda K., Sakurai Takashi, Stewart Jean K., Yimlamai Dean, Unger Christine, Zehetmeier Carol, Lain Blanca, Torella Claudia, Henning Stefan W., Beste Gerald, Scroggins Bradley T., Neckers Len, Ilag Leodevico L., Jay Daniel G.. Nature Cell Biology.2004;6(6). CrossRef
- Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling Weissmueller Susann, Manchado Eusebio, Saborowski Michael, Morris John P., Wagenblast Elvin, Davis Carrie A., Moon Sung-Hwan, Pfister Neil T., Tschaharganeh Darjus F., Kitzing Thomas, Aust Daniela, Markert Elke K., Wu Jianmin, Grimmond Sean M., Pilarsky Christian, Prives Carol, Biankin Andrew V., Lowe Scott W.. Cell.2014;157(2). CrossRef
- Effect of Mutant p53 Proteins on Glycolysis and Mitochondrial Metabolism Eriksson Matilda, Ambroise Gorbatchev, Ouchida Amanda Tomie, Lima Queiroz Andre, Smith Dominique, Gimenez-Cassina Alfredo, Iwanicki Marcin P., Muller Patricia A., Norberg Erik, Vakifahmetoglu-Norberg Helin. Molecular and Cellular Biology.2017;37(24). CrossRef
- p53 regulates mitochondrial respiration Matoba Satoaki, Kang Ju-Gyeong, Patino Willmar D., Wragg Andrew, Boehm Manfred, Gavrilova Oksana, Hurley Paula J., Bunz Fred, Hwang Paul M.. Science (New York, N.Y.).2006;312(5780). CrossRef
- Gain-of-function mutant p53 in cancer progression and therapy Zhang Cen, Liu Juan, Xu Dandan, Zhang Tianliang, Hu Wenwei, Feng Zhaohui. Journal of Molecular Cell Biology.2020;12(9). CrossRef
- The interplay between cell signalling and the mevalonate pathway in cancer Mullen Peter J., Yu Rosemary, Longo Joseph, Archer Michael C., Penn Linda Z.. Nature Reviews. Cancer.2016;16(11). CrossRef
- Metabolic control of YAP and TAZ by the mevalonate pathway Sorrentino Giovanni, Ruggeri Naomi, Specchia Valeria, Cordenonsi Michelangelo, Mano Miguel, Dupont Sirio, Manfrin Andrea, Ingallina Eleonora, Sommaggio Roberta, Piazza Silvano, Rosato Antonio, Piccolo Stefano, Del Sal Giannino. Nature Cell Biology.2014;16(4). CrossRef
- The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways Feng Zhaohui, Hu Wenwei, Stanchina Elisa, Teresky Angelika K., Jin Shengkan, Lowe Scott, Levine Arnold J.. Cancer Research.2007;67(7). CrossRef
- Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation Zhou Ge, Wang Jiping, Zhao Mei, Xie Tong-Xin, Tanaka Noriaki, Sano Daisuke, Patel Ameeta A., Ward Alexandra M., Sandulache Vlad C., Jasser Samar A., Skinner Heath D., Fitzgerald Alison Lea, Osman Abdullah A., Wei Yongkun, Xia Xuefeng, Songyang Zhou, Mills Gordon B., Hung Mien-Chie, Caulin Carlos, Liang Jiyong, Myers Jeffrey N.. Molecular Cell.2014;54(6). CrossRef
- TIGAR, a p53-inducible regulator of glycolysis and apoptosis K Bensaad, A Tsuruta, Ma Selak, Mn Vidal, K Nakano, R Bartrons, E Gottlieb, Kh Vousden. Cell.2006;126(1). CrossRef
- TIGAR is required for efficient intestinal regeneration and tumorigenesis Cheung Eric C., Athineos Dimitris, Lee Pearl, Ridgway Rachel A., Lambie Wendy, Nixon Colin, Strathdee Douglas, Blyth Karen, Sansom Owen J., Vousden Karen H.. Developmental Cell.2013;25(5). CrossRef
- Heme oxygenase-1 determines the differential response of breast cancer and normal cells to piperlongumine Lee Ha-Na, Jin Hyeon-Ok, Park Jin-Ah, Kim Jin-Hee, Kim Ji-Young, Kim BoRa, Kim Wonki, Hong Sung-Eun, Lee Yun-Han, Chang Yoon Hwan, Hong Seok-Il, Hong Young Jun, Park In-Chul, Surh Young-Joon, Lee Jin Kyung. Molecules and Cells.2015;38(4). CrossRef
- Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ Escoll M., Gargini R., Cuadrado A., Anton I. M., Wandosell F.. Oncogene.2017;36(25). CrossRef
- A Gain-of-Function p53-Mutant Oncogene Promotes Cell Fate Plasticity and Myeloid Leukemia through the Pluripotency Factor FOXH1 Loizou Evangelia, Banito Ana, Livshits Geulah, Ho Yu-Jui, Koche Richard P., Sánchez-Rivera Francisco J., Mayle Allison, Chen Chi-Chao, Kinalis Savvas, Bagger Frederik O., Kastenhuber Edward R., Durham Benjamin H., Lowe Scott W.. Cancer Discovery.2019;9(7). CrossRef
- An N6-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells Uddin Mohammad B., Roy Kartik R., Hosain Salman B., Khiste Sachin K., Hill Ronald A., Jois Seetharama D., Zhao Yunfeng, Tackett Alan J., Liu Yong-Yu. Biochemical Pharmacology.2019;160. CrossRef
- TP53 status predicts long-term survival in locally advanced breast cancer after primary chemotherapy Eikesdal Hans P., Knappskog Stian, Aas Turid, Lønning Per E.. Acta Oncologica (Stockholm, Sweden).2014;53(10). CrossRef
- MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function Donzelli S., Fontemaggi G., Fazi F., Di Agostino S., Padula F., Biagioni F., Muti P., Strano S., Blandino G.. Cell Death & Differentiation.2012;19(6). CrossRef
- Intrinsic adriamycin resistance in p53-mutated breast cancer is related to the miR-30c/FANCF/REV1-mediated DNA damage response Lin Shu, Yu Lifeng, Song Xinyue, Bi Jia, Jiang Longyang, Wang Yan, He Miao, Xiao Qinghuan, Sun Mingli, Olopade Olufunmilayo I., Zhao Lin, Wei Minjie. Cell Death & Disease.2019;10(9). CrossRef
- Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression Wang W., Cheng B., Miao L., Mei Y., Wu M.. Cell Death & Disease.2013;4. CrossRef
- EFNB2 facilitates cell proliferation, migration, and invasion in pancreatic ductal adenocarcinoma via the p53/p21 pathway and EMT Zhu Feng, Dai Shang-Nan, Xu Da-Lai, Hou Chao-Qun, Liu Tong-Tai, Chen Qiu-Yang, Wu Jun-Li, Miao Yi. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie.2020;125. CrossRef
- DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53 Alam S. K., Yadav V. K., Bajaj S., Datta A., Dutta S. K., Bhattacharyya M., Bhattacharya S., Debnath S., Roy S., Boardman L. A., Smyrk T. C., Molina J. R., Chakrabarti S., Chowdhury S., Mukhopadhyay D., Roychoudhury S.. Cell Death and Differentiation.2016;23(4). CrossRef
- p53 and Ca(2+) signaling from the endoplasmic reticulum: partners in anti-cancer therapies Bittremieux Mart, Bultynck Geert. Oncoscience.2015;2(3). CrossRef
- p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner Giorgi Carlotta, Bonora Massimo, Sorrentino Giovanni, Missiroli Sonia, Poletti Federica, Suski Jan M., Galindo Ramirez Fabian, Rizzuto Rosario, Di Virgilio Francesco, Zito Ester, Pandolfi Pier Paolo, Wieckowski Mariusz R., Mammano Fabio, Del Sal Giannino, Pinton Paolo. Proceedings of the National Academy of Sciences.2015;112(6). CrossRef
- Mutant p53 drives cancer chemotherapy resistance due to loss of function on activating transcription of PUMA Huang Yuan, Liu Nannan, Liu Jing, Liu Yeying, Zhang Chuchu, Long Shuaiyu, Luo Guang, Zhang Lingling, Zhang Yingjie. Cell Cycle (Georgetown, Tex.).2019;18(24). CrossRef
- Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity Xu J., Wang J., Hu Y., Qian J., Xu B., Chen H., Zou W., Fang J.-Y.. Cell Death & Disease.2014;5(3). CrossRef
- Cancer. p53, guardian of the genome Lane D. P.. Nature.1992;358(6381). CrossRef
- The many faces of p53: something for everyone Levine Arnold J.. Journal of Molecular Cell Biology.2019;11(7). CrossRef
- Mutant p53 promotes tumor progression and metastasis by the endoplasmic reticulum UDPase ENTPD5 Vogiatzi Fotini, Brandt Dominique T., Schneikert Jean, Fuchs Jeannette, Grikscheit Katharina, Wanzel Michael, Pavlakis Evangelos, Charles Joël P., Timofeev Oleg, Nist Andrea, Mernberger Marco, Kantelhardt Eva J., Siebolts Udo, Bartel Frank, Jacob Ralf, Rath Ariane, Moll Roland, Grosse Robert, Stiewe Thorsten. Proceedings of the National Academy of Sciences.2016;113(52). CrossRef
- Role of p53 in Cell Death and Human Cancers Ozaki Toshinori, Nakagawara Akira. Cancers.2011;3(1). CrossRef
- TP53 mutations in human cancers: origins, consequences, and clinical use Olivier Magali, Hollstein Monica, Hainaut Pierre. Cold Spring Harbor Perspectives in Biology.2010;2(1). CrossRef
- Apoptosis in cancer therapy: crossing the threshold Fisher D. E.. Cell.1994;78(4). CrossRef
- In vivo activation of the p53 pathway by small-molecule antagonists of MDM2 Vassilev Lyubomir T., Vu Binh T., Graves Bradford, Carvajal Daisy, Podlaski Frank, Filipovic Zoran, Kong Norman, Kammlott Ursula, Lukacs Christine, Klein Christian, Fotouhi Nader, Liu Emily A.. Science (New York, N.Y.).2004;303(5659). CrossRef
- MI-219-zinc combination: a new paradigm in MDM2 inhibitor-based therapy Azmi A. S., Philip P. A., Beck F. W. J., Wang Z., Banerjee S., Wang S., Yang D., Sarkar F. H., Mohammad R. M.. Oncogene.2011;30(1). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details