Assessing Mutations in Treatment Response-related Genes in Egyptian Patients with Non-small Cell Lung Cancer
Download
Abstract
Background: Epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene homolog (K-RAS) are the most common driver genes in patients with non-small cell lung cancer adenocarcinomas (NSCLC/ADC), which affects treatment. Therefore, this study determines the frequency and patterns of EGFR and K-RAS mutations in Egyptian patients with NSCLC/ADC and correlates them with clinicopathological features.
Methods: A retrospective analysis of 139 patients diagnosed with NSCLC/ADC and screened for EGFR and K-RAS mutations was conducted; further evaluating clinicopathological characteristics and mutational status.
Results: This study included 101 males and 38 females with a median age of 57.7 ± 10.5 years. EGFR mutations were detected in 22.3% (12.2% in exon 19, 8.6% in exon 21, and 1.4% in exon 18) and K-RAS mutations in 17.3% (15.8% in codon 12 and 1.4% in codon 13), whereas combined mutations were detected in nine patients (6.5%). Furthermore, EGFR mutations were non-significantly more common in females and nonsmokers, contradicting K-RAS mutations, which were more common in males and smokers.
Conclusion: EGFR and K-RAS mutations are common in Egyptian patients with NSCLC/ADC (National Cancer Institute experience). Their incidences were between the Asian Pacific and Europeans. Also, their mutations led to dysregulation in tyrosine kinase activity, which correlates with the late disease stage and poor progression. Therefore, analyzing them should be done to determine a better treatment method and predict survival outcomes.
Introduction
For the past two decades, targeting mutations in the kinase domain of the epidermal growth factor receptor (EGFR) gene has been an important approach used to treat non-small cell lung cancer (NSCLC) tumors. Unfortunately, according to global cancer incidence, mortality, and prevalence (GLOBOCAN) 2020, lung cancer is the leading cause of cancer-related death, with an estimated 1.8 million deaths [1].
The EGFR receptor is a transmembrane glycoprotein receptor, and its gene is located on chromosome 7’s short arm (7p11.2) that encodes the 170 kDa Type I transmembrane growth factor receptor with tyrosine kinase (TK) activity [2]. Additionally, EGFR belongs to the human epidermal growth factor receptor/erbB family of receptor TKs, which received great attention due to its strong association with malignant transformation and cellular proliferation. Homodimerization and/or heterodimerization in response to ligand-binding activate the TK, causing autophosphorylation of the receptor’s cytoplasmic domain. This allows interaction with other molecules, activating three major downstream signaling pathways. These pathways are important in maintaining and growing normal cells [3-6]. Therefore EGFR is a vital gene responsible for cell activity; its mutation is accompanied by a dysregulation in EGFR TK activity, which is closely correlated with the late disease stage and poor progression of NSCLC [7,8]. The most common mutations in the EGFR gene are mostly in the four exons (18–21) that are clustered around the TK domain [9]. They include point mutation G719X (G719A, G719C, G719S) in exon 18, in-frame deletions and in-frame insertions in exon 19, point mutation T790M and insertions in exon 20, and point mutations (L858R and L861Q) in exon 21. However, of these mutations, only two types account for about 85% of the EGFR mutations: deletions in exon 19 and L858R point mutation in exon 21 [10].
In 2004, EGFR mutations were found to be sensitive to targeted therapies called tyrosine kinase inhibitors (TKIs), which were the first molecular changes in NSCLC cases. Also, chimeric monoclonal antibodies (panitumumab and cetuximab) and TKIs (gefitinib, erlotinib, and afatinib) are among these developed strategies that target EGFR [11] [12] They were chosen to compete with ATP (adenosine triphosphate) binding site inhibitors at the active site of EGFR kinase, therefore preventing and blocking vital EGFR pathways [13,14].
The TKIs of the first-generation (erlotinib, gefitinib, and icotinib) induce reversible ATP-binding site blockade and halt the downstream signaling pathway. Almost all patients who received this generation developed resistance. The T790M point mutation was among the most common mutations in patients who developed resistance to first-generation TKIs. Second-generation compounds (afatinib and dacomitinib) formed irreversible covalent bonds with all homo- and hetero-dimers of the ErbB family receptors and blocked the transphosphorylation to inhibit signaling. Thus, having a greater effect to overcome or delay the resistance of first-generation TKIs, but it also acquired resistance. The third and most recent generation (osimertinib, rociletinib, olmutinib, and lazertinib) is a novel treatment due to its ability to bind to the T790M EGFR-mutant receptor. However, patients who received third-generation TKIs developed resistance. Other factors, such as small cell lung cancer transformation and downstream gene mutations [Kristen rat sarcoma viral oncogene homolog (K-RAS) and v-raf murine sarcoma viral oncology homolog B (BRAF)] were discovered in patients who developed resistance [12] [15]. The RAS family proteins are GTPase proteins that switch various cellular activities. This family includes K-RAS, H-RAS, and N-RAS genes [16], and its mutation is usually associated with TKI resistance, as mentioned above. Therefore, coexisting driver mutations in the same tumor affect the therapeutic outcome and survival rate of patients with NSCLC, whether treated by chemotherapy or targeted therapies [17]. Codons 12/13 K-RAS mutations were described in approximately 20% of lung adenocarcinomas and are associated with tobacco consumption [18].
The prevalence of EGFR mutations varied greatly across ethnic backgrounds and geographical locations due to differences in patient demographics, study designs, assays used, number of sequenced exons, tumor source, and eligibility criteria for trial enrolment [19]. In 2015 study, 50 lung tissue biopsies from Egyptian patients were screened for EGFR mutations in which, deletion in exon 19 was the only detected mutation [20]. Recently, in a 2020 study, EGFR mutation was detected in 15/34 (44.1% of the tumors) using EGFR XL Strip Assay kit [21] and in 2022 anther study was published where EGFR mutations were detected in 40.8% of NSCLC Egyptian patients [22]. This study determines the frequency and pattern (s) of EGFR and K-RAS mutations in NSCLC adenocarcinoma (ADC) cases from Egypt. Furthermore, it correlates the detected mutations to the patients’ relevant clinicopathological data.
Materials and Methods
Subjects
A retrospective cohort study was conducted in which 139 patients with NSCLC/ADC were recruited from the National Cancer Institute (NCI) data clinics between 2012 and 2015. Ethical approval was granted by the Institutional Review Board of the Egyptian NCI (approval no.2010011003.3). Notably, this study was conducted following the Helsinki Declaration.
Sample preparation and DNA Extraction
From each specimen, a representative hematoxylin and eosin stained slide was prepared and reviewed by a pathologist to ensure the presence of adequate representative tumor cells in the section before extraction. DNA was then extracted and purified from the selected formalin-fixed, paraffin-embedded (FFPE) tissue blocks (each of the five sections is 4–5 micron thick) using the QIAamp DNA FFPE tissue kit (Qiagen, Germany) (catalog number: 56404) according to manufacturers’ protocols.
EGFR mutational analysis
EGFR mutations were detected using the Therascreen EGFR RGQ PCR kit (Qiagen, Germany) (catalog number: 870111) on Rotor-Gene Q MDx 5plex HRM instrument (Germany) according to manufacturers’ protocol. This ready-to-use kit detects somatic mutations in the EGFR gene by PCR. In addition, it detects actionable somatic mutations against a background of wild-type (WT) genomic DNA using ARMS (Amplified Refractory Mutation System) and Scorpion techniques. The detected mutations were exon 19 deletions, T790M, L858R, L861Q, G719X, and S768I, and three insertions in exon 20. Mutation types are listed in Table 1.
| Mutation | Exon | Base change | COSMIC ID |
| T790M | 20 | 2369C>T | 6240 |
| L858R | 21 | 2573T>G | 6224 |
| L861Q | 21 | 2582T>A | 6213 |
| S768I | 20 | 2303G>T | 6241 |
| G719A | 18 | 2156G>C | 6239 |
| G719S | 18 | 2155G>A | 6252 |
| G719C | 18 | 2155G>T | 6253 |
| Insertions | 20 | 2307_2308ins9 | 12376 |
| 2319_2320insCAC | 12377 | ||
| 2310_2311insGGT | 12378 | ||
| Deletion | 19 | 2235_2249del15 | 6223 |
| 2235_2252>AAT(complex) | 13551 | ||
| 2236_2253del18 | 12728 | ||
| 2237_2251del15 | 12678 | ||
| 2237_2254del18 | 12367 | ||
| 2237_2255>T (complex) | 12384 | ||
| 2236_2250del15 | 6225 | ||
| 2238_2255del18 | 6220 | ||
| 2238_2248>GC (complex) | 12422 | ||
| 2238_2252>GCA(complex) | 12419 | ||
| 2239_2247del9 | 6218 | ||
| 2239_2253del15 | 6254 | ||
| 2239_2256del18 | 6255 | ||
| 2239_2248TTAAGAGAAGC (complex) | 12382 | ||
| 2239_2258>CA (complex) | 12387 | ||
| 2240_2251del12 | 6210 | ||
| 2240_2257del18 | 12370 | ||
| 2240_2254del15 | 12369 | ||
| 2239_2251>C (complex) | 12383 |
The test was performed twice; the first sample assessed the total amplified DNA, ensuring that each sample contained a sufficient amount of DNA. The second is to determine the presence or absence of EGFR mutations and the type of mutation (s).
K-RAS mutational analysis
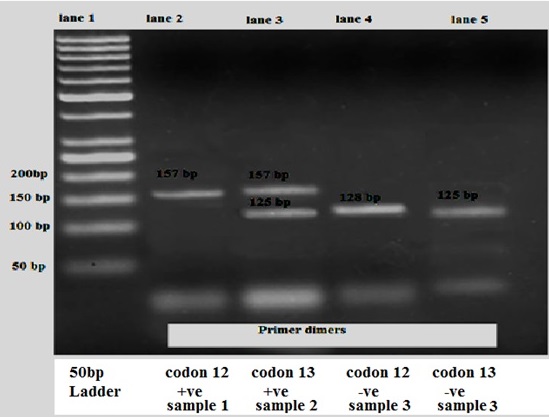
According to a method by Hadija (2007) [23], each sample underwent PCR-RFLP (PCR with restriction fragmentation length polymorphism) analysis for PCR amplification of codons 12 and 13 of the K-RAS gene. The PCR products were then digested using restriction enzymes Bst NI for codon 12 mutations and Bgl I for codon 13 mutations. The test was conducted in four stages: first, PCR and digestion; then second, PCR and digestion. For UV visualization, the second digestion’s final codon product was then separated on a 2% agarose gel stained with ethidium bromide. Also, product sizes were estimated by comparing them to the 50 bp DNA ladder. The 157 bp and 128 bp fragments represent mutation at codon 12 and WT, respectively. For codon 13, the 157 bp and 125 bp fragments represent mutation and WT, respectively [23].
Statistical analysis
Statistical analysis was performed using the Statistical Package for Social Sciences v.24. Numerical data were summarized using means and standard deviations (SD) or medians and ranges. Categorical variables were summarized as numbers and percentages; differences were analyzed with the X2 (chi square) and Fisher’s exact tests. Finally, P values of ≤0.05 were considered significant.
Results
Of the 139 patients enrolled in this study, 101 (72.7%) were males, and 38 (27.3%) were females. The mean age was 57.7 years (26–81). Fifty-two cases (37.4%) were nonsmokers, however, 87 (62.6%) were current and former smokers. All included patients were diagnosed with adenocarcinoma, and most cases were in stage III (26 patients: 19.8%) and stage IV (99 patients: 75.6%). One hundred and twenty-two patients received chemotherapy (92.4%), whereas surgery was performed in 20 patients (15.3%), and radiotherapy (RTH) was given to four patients (3%). The main demographic and clinical data of the patients included in this study are illustrated in Table 2.
| Characteristics | Number (Percentage) | |
| Gender | Male | 101 (72) |
| Female | 38 (27.3) | |
| Age (years) | ≤59 | 69 (49.6) |
| Mean ± SD (range) | >59 | 70 (50.4) |
| male | 58.4±10.5 (30-81) | |
| female | 55.8±10.4 (26-75) | |
| Total | 57.7±10.5 (26-81) | |
| Smoking | nonsmokers | 52 (37.4) |
| Smokers | 87 (62.6) | |
| Tumor size (cm) | mean± SD | 6.3±2.7 |
| range | (1-11) | |
| Performance status | 1 | 98 (76) |
| 2 | 24 (18.6) | |
| 3 | 7 (5.4) | |
| Pathology | adenocarcinoma | 139(100) |
| Tumor grade | 1 | 5 (4.4) |
| 2 | 67 (58.8) | |
| 3 | 42 (36.8) | |
| Tumor stage | I | 3 (2.3) |
| II | 3 (2.3) | |
| III | 26 (19.8) | |
| SD; standard deviation | IV | 99 (75.6) |
Furthermore, EGFR mutations were detected in 31/139 cases (22.3%), and they were non-significantly more frequent in females than males (31.6% vs. 18.8%) and nonsmokers compared with smokers (28.8% vs. 18.4%) (Table 3).
| Clinicopathological feature | EGFR | K-RAS | |||||||||
| Wild | Mutant | p value | Wild | Mutant | p value | ||||||
| No | % | No | % | ≤ | No | % | No | % | ≤ | ||
| Age | ≤59 | 52 | 75.4 | 17 | 24.6 | NS | 60 | 87 | 9 | 13 | NS |
| (years) | >59 | 56 | 80 | 14 | 20 | NS | 55 | 78.6 | 15 | 21.4 | NS |
| Gender | male | 82 | 81.2 | 19 | 18.8 | NS | 83 | 82.2 | 18 | 17.8 | NS |
| female | 26 | 68.4 | 12 | 31.6 | NS | 32 | 84.2 | 6 | 15.8 | NS | |
| Smoking | nonsmokers | 37 | 71.2 | 15 | 28.8 | NS | 44 | 84.6 | 8 | 15.4 | NS |
| smoker | 71 | 81.6 | 16 | 18.4 | NS | 71 | 81.6 | 16 | 18.4 | NS | |
| Performance status | 1 | 77 | 78.6 | 21 | 21.4 | NS | 84 | 85.7 | 14 | 14.3 | NS |
| 2 | 18 | 75 | 6 | 25 | NS | 18 | 75 | 6 | 25 | NS | |
| 3 | 6 | 85.7 | 1 | 14.3 | NS | 6 | 85.7 | 1 | 14.3 | NS | |
| Tumor | Low grade | 64 | 88.9 | 8 | 11.1 | NS | 62 | 86.1 | 10 | 13.9 | NS |
| Grade | high grade | 19 | 45.2 | 23 | 54.8 | <0.001 | 30 | 71.4 | 12 | 28.6 | <0.05 |
The most common detected mutations were deletion in exon 19 followed by exon 21 point mutation, accounting for 12.2% and 8.6% of all 139 samples, respectively. However, only two cases had a mutation in exon 18 G719X (1.4%) (Figures 1 and 2).
Figure 1. Frequency of EGFR and Its Mutational Pattern.
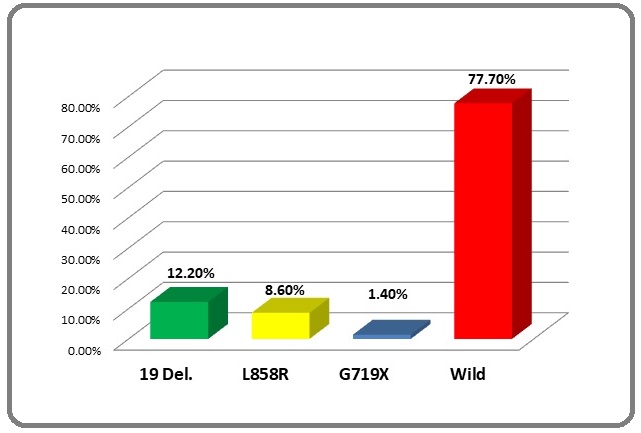
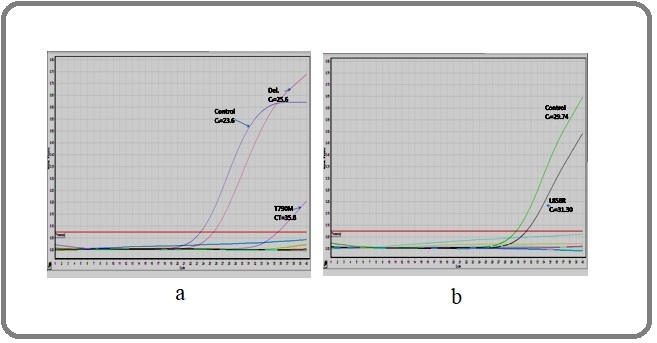
Figure 2. EGFR Amplification Curves. In Figure 2a, three curves were detected: control CT = 23.6, T790M CT = 35.58 (∆CT = 11.98 not within the cutoff range; a cutoff range of ∆CT ≤ 7.40), and Del 19 CT = 25.6 (∆CT = 2 within the cutoffrange; a cutoff range of ∆CT ≤ 8.90); therefore, this case was positive for deletion in exon 19. Meanwhile, two curves were detected in Figure 2b: control CT= 29.74 and L858R CT = 31.3 ∆CT = 1.59 within the cutoff range (cutoff range T of ∆CT ≤ 8.90). Therefore this case was positive for exon 21.
K-RAS mutations were found in 24/139 (17.3%) patients and were non-significantly more frequent in smokers compared with nonsmokers (18.4% vs. 15.4%) and males rather than females (17.8% vs. 15.8%) (Table 3). The most common K-RAS mutations were found in codon 12 (22 patients, 15.8%), whereas codon 13 was found in two patients (1.4%) (Figures 3 and 4)
Figure 3. Frequency of K-RAS and Its Mutational Pattern.
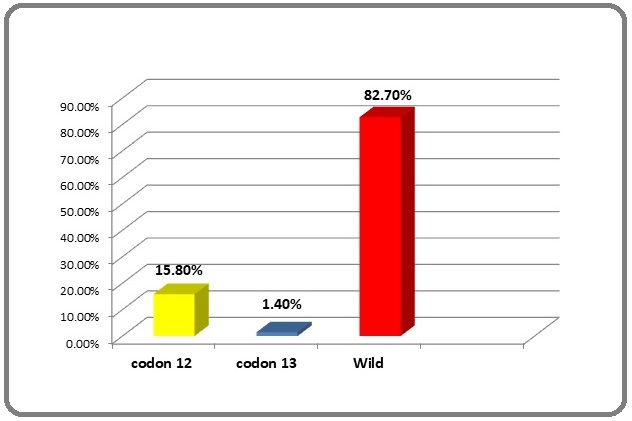
Figure 4. K-RAS Mutation Bands after RFLP Analysis. Lane 1: 50 bp ladder. Lane 2: 157 bp band, representing positive mutation for codon 12 (sample 1). Lane 3: 157 bp band, representing positive mutation for codon 13 allele and 125 bp band, representing WT for codon 13 allele (sample 2) (heterozygous sample). Lane 4: 128 bp band, representing WT for codon 12 (sample 3). Lane 5: 125 bp band, representing WT for codon 13 (sample 3).
Nine patients (6.5%) had mutations in both genes (EGFR and K-RAS), whereas 93 (66.9%) were WT for both genes.
High grades showed a significantly higher proportion of all clinicopathological factors with EGFR and K-RAS mutations (Table 3).
Discussion
This study detected EGFR gene mutations in 31 patients with NSCLC/ADC (22.3%), whereas K-RAS mutations were found in 24 patients (17.3%). Previous studies by Benbrahim (2018) [24] and Tfayli [19] reported that EGFR mutation frequency in patients in the Middle East and Africa is higher than that in white populations but still lower than that reported in Asia. These results agree with those in this study, which showed that the mutational incidence of EGFR was higher than that of the Asian pacific regions but lower than the European incidences [25-27].
Most of the last decade studies showed that Asia- Pacific regions (China, Hong Kong, Japan, Malaysia, Philippines, Korea, Singapore, Taiwan, Thailand, Vietnam, and India) had the highest EGFR gene mutation frequency [25] [28,29] however, K-RAS mutation incidences in these regions were low [30-32].
Conversely, in European regions (Czech Republic, Finland, France, Germany, Greece, Italy, Lithuania, the Netherlands, Norway, Portugal, Russia, Slovakia, Spain, Sweden, Turkey, and United Kingdom), EGFR mutation frequencies were lower than Asia’s incidences and those reported in this study [33,34]. Meanwhile, K-RAS mutational frequencies in the European regions were much higher than in the Asian regions [18] [32] [34,35]. Furthermore, in our study, the prevalence of K-RAS mutation was also between the European and Asian regions.
In this study, EGFR and K-RAS combined gene mutations were found in nine patients (6.5%), and this incidence is higher than that observed in a Vietnamese study, in which combined mutations were 1.7% [36].
The reported EGFR mutational incidence in this study was close to a Brazilian study conducted in 2019 where EGFR mutation was found in 22.7% of 444 Brazilian patients, whereas K-RAS mutation was in 20.4% [18].
Also, the incidence in our study agreed with that in a study conducted in Morocco in 2013 [37].
Although the prevalence of EGFR incidence rate in this study agreed with the Middle East region rate (2.9%–28.7%) [20] [24] [37], it was lower than that demonstrated in Iraq (27.53%) [38]. Also, it was lower in the four Gulf regions (United Arab Emirates, Kuwait, Oman, and Qatar) in 2020 (36.9%) [39], but higher than that detected in the Saudi populations, Lebanese populations, and the multicentre study done in Lebanon, Jordan, and Iraq [40-44].
ARMS is considered one of the most sensitive techniques as it could detect mutation as low as 0.1%–1% [45]. Using ARMS [46] and Scorpion [47] techniques, the frame deletion of exon 19 and point mutations in exon 21 in our study were the most common EGFR-detected mutations (12.2% and 8.2%, respectively), coinciding with most of the published data [28] [35] [44] [48,49]. In contrast, the G719X point mutation was only detected in two cases (1.4%), which is lower than that reported by Errihani (2013) [37] and Jazeih (2013 and 2015) [50,51]. For K-RAS mutations, codon 12 was the most common (15.8%) in our study, which agrees with previous findings [31,32] [52].
Detecting different mutational patterns of EGFR is crucial to consider during EGFR analysis, which contributes to variations in response to treatment, prognosis, and survival rates according to the different EGFR mutations. Better prognosis and long survival were associated with the mutation in exons 19 and 21, whereas poor prognosis was correlated with a mutation in exon 20 [53,54]. In contrast, the response rate to TKIs was improved in exons 19 and 21 mutations; exon 20 showed resistance to the therapy [38][55]. Also, exon 20 of the EGFR gene and other downstream gene mutations showed resistance. K-RAS gene mutations are usually associated with TKI resistance [12] [15] [17].
In most studies, EGFR mutations were statistically significant with females and nonsmokers [19] [36] [56]. However, in our study, EGFR mutations were non-significantly more common in females and nonsmokers, which could be attributed to the fewer number of female patients compared with males (38 females vs. 101 males) and the fewer number of smokers compared with nonsmokers (52 nonsmokers vs. 87 smokers), coinciding with the studies of Unal (2013) [55] in Turkey and Ramadhan (2021) [38] in Iraq.
However, Dang (2020) [36] claimed that K-RAS mutation frequency was significantly higher in males than female patients and smokers than nonsmokers, which agreed with our study.
Conclusively, EGFR mutations were found in 22.3% of Egyptian patients with NSCLC/ADC and K-RAS in 17.3%, and the most frequent EGFR mutations detected in this study were in exons 19 and 21. Also, the prevalence of EGFR mutations was non-significantly higher in females than males and nonsmokers than smokers. Conversely, the K-RAS mutation was more common in males than females and smokers than nonsmokers. Furthermore, these frequencies were between the white and Asian populations, and they must be considered since they affect treatment and help predict the response of patients and their survival rates.
References
- Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Sung H, Ferlay J, Rebecca L, et al . Ca Cancer J Clin.2021;0:1-41.
- Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial Rosell Rafael, Carcereny Enric, Gervais Radj, Vergnenegre Alain, Massuti Bartomeu, Felip Enriqueta, Palmero Ramon, Garcia-Gomez Ramon, Pallares Cinta, Sanchez Jose Miguel, Porta Rut, Cobo Manuel, Garrido Pilar, Longo Flavia, Moran Teresa, Insa Amelia, De Marinis Filippo, Corre Romain, Bover Isabel, Illiano Alfonso, Dansin Eric, Castro Javier, Milella Michele, Reguart Noemi, Altavilla Giuseppe, Jimenez Ulpiano, Provencio Mariano, Moreno Miguel Angel, Terrasa Josefa, Muñoz-Langa Jose, Valdivia Javier, Isla Dolores, Domine Manuel, Molinier Olivier, Mazieres Julien, Baize Nathalie, Garcia-Campelo Rosario, Robinet Gilles, Rodriguez-Abreu Delvys, Lopez-Vivanco Guillermo, Gebbia Vittorio, Ferrera-Delgado Lioba, Bombaron Pierre, Bernabe Reyes, Bearz Alessandra, Artal Angel, Cortesi Enrico, Rolfo Christian, Sanchez-Ronco Maria, Drozdowskyj Ana, Queralt Cristina, Aguirre Itziar, Ramirez Jose Luis, Sanchez Jose Javier, Molina Miguel Angel, Taron Miquel, Paz-Ares Luis. The Lancet. Oncology.2012;13(3). CrossRef
- EGFR antagonists in cancer treatment Ciardiello Fortunato, Tortora Giampaolo. The New England Journal of Medicine.2008;358(11). CrossRef
- Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer Eck Michael J., Yun Cai-Hong. Biochimica Et Biophysica Acta.2010;1804(3). CrossRef
- EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications Yasuda Hiroyuki, Kobayashi Susumu, Costa Daniel B.. The Lancet. Oncology.2012;13(1). CrossRef
- Mechanisms of resistance to EGFR tyrosine kinase inhibitors Huang Lihua, Fu Liwu. Acta Pharmaceutica Sinica. B.2015;5(5). CrossRef
- Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology Jänne Pasi A., Engelman Jeffrey A., Johnson Bruce E.. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2005;23(14). CrossRef
- EGFR mutations and lung cancer Cunha Santos Gilda, Shepherd Frances A., Tsao Ming Sound. Annual Review of Pathology.2011;6. CrossRef
- Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers Yu Helena A., Arcila Maria E., Rekhtman Natasha, Sima Camelia S., Zakowski Maureen F., Pao William, Kris Mark G., Miller Vincent A., Ladanyi Marc, Riely Gregory J.. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2013;19(8). CrossRef
- Pathogenesis of lung cancer signalling pathways: roadmap for therapies Brambilla E., Gazdar A.. The European Respiratory Journal.2009;33(6). CrossRef
- Epidermal growth factor receptor inhibitors in the treatment of non-small-cell lung cancer Giaccone Giuseppe. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2005;23(14). CrossRef
- Epidermal growth factor receptor (EGFR): A rising star in the era of precision medicine of lung cancer Liu Xiaomin, Wang Ping, Zhang Caiyan, Ma Zhongliang. Oncotarget.2017;8(30). CrossRef
- EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib Pao William, Miller Vincent, Zakowski Maureen, Doherty Jennifer, Politi Katerina, Sarkaria Inderpal, Singh Bhuvanesh, Heelan Robert, Rusch Valerie, Fulton Lucinda, Mardis Elaine, Kupfer Doris, Wilson Richard, Kris Mark, Varmus Harold. Proceedings of the National Academy of Sciences of the United States of America.2004;101(36). CrossRef
- Protein kinase inhibitors: contributions from structure to clinical compounds Johnson Louise N.. Quarterly Reviews of Biophysics.2009;42(1). CrossRef
- Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer Leonetti Alessandro, Sharma Sugandhi, Minari Roberta, Perego Paola, Giovannetti Elisa, Tiseo Marcello. British Journal of Cancer.2019;121(9). CrossRef
- KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib Pao William, Wang Theresa Y., Riely Gregory J., Miller Vincent A., Pan Qiulu, Ladanyi Marc, Zakowski Maureen F., Heelan Robert T., Kris Mark G., Varmus Harold E.. PLoS medicine.2005;2(1). CrossRef
- Concomitant ALK/KRAS and ALK/EGFR mutations in non-small cell lung cancer: different profile of response to target therapies Marino FZ, Ronchi A, Accardo M, Franco R. Trans Cancer Res.2017;6:S457-60.
- Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations Leal Letícia Ferro, Paula Flávia Escremim, De Marchi Pedro, Souza Viana Luciano, Pinto Gustavo Dix Junqueira, Carlos Carolina Dias, Berardinelli Gustavo Noriz, Miziara José Elias, Silva Carlos Maciel, Silva Eduardo Caetano Albino, Pereira Rui, Oliveira Marco Antonio, Scapulatempo-Neto Cristovam, Reis Rui Manuel. Scientific Reports.2019;9(1). CrossRef
- Prevalence of the epidermal growth factor receptor mutations in lung adenocarcinoma patients from the Middle East region Tfayli Arafat Hussein, Fakhri Ghina Bassam, Al Assaad Majd Sassine. Annals of Thoracic Medicine.2019;14(3). CrossRef
- Nonenriched PCR Versus Mutant-Enriched PCR in Detecting Selected Epidermal Growth Factor Receptor Gene Mutations Among Nonsmall-Cell Lung Cancer Patients Zaki Moyassar Ahmad, Ramadan Ragaa Abd El Kader, Mahmoud Mahmoud Ibrahim, El-Kaffash Dalal Mohammed, Assaad Rami Samir. Genetic Testing and Molecular Biomarkers.2015;19(8). CrossRef
- Mutation of EGFR in non-small cell lung cancer, a regional study in Upper Egypt Eid SS, Ahmed AR, Abdelgawad MI, et al . Cancer Research Journal.2020;8:1-7.
- Mutation patterns of epidermal growth factor receptor gene in non-small cell lung cancer among Egyptian patients Elmetnawy Wafaa H., Qenawi Mona, Sabet Salwa, Bassiony Heba. Egyptian Journal of Basic and Applied Sciences.2022;9(1). CrossRef
- K-Ras and Dpc4 mutations in chronic pancreatitis: case series. Hadžija MP, Korolija M, Razumović JJ, et al . Croat Med J.2007;48:218-24.
- EGFR mutation frequency in Middle East and African non-small cell lung cancer patients: a systematic review and meta-analysis Benbrahim Zineb, Antonia Teresita, Mellas Nawfel. BMC cancer.2018;18(1). CrossRef
- EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Midha A, Dearden S, McCormack R. Am J Cancer Res.2015;5:2892-911.
- The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis Zhang Yue-Lun, Yuan Jin-Qiu, Wang Kai-Feng, Fu Xiao-Hong, Han Xiao-Ran, Threapleton Diane, Yang Zu-Yao, Mao Chen, Tang Jin-Ling. Oncotarget.2016;7(48). CrossRef
- Worldwide Frequency of Commonly Detected EGFR Mutations Graham Rondell P., Treece Amanda L., Lindeman Neal I., Vasalos Patricia, Shan Mu, Jennings Lawrence J., Rimm David L.. Archives of Pathology & Laboratory Medicine.2018;142(2). CrossRef
- Clinical significance of EGFR mutation types in lung adenocarcinoma: A multi-centre Korean study Yoon Hee-Young, Ryu Jeong-Seon, Sim Yun Su, Kim Dojin, Lee Sung Yong, Choi Juwhan, Park Sojung, Ryu Yon Ju, Lee Jin Hwa, Chang Jung Hyun. PLOS ONE.2020;15(2). CrossRef
- Mutation profile of non-small cell lung cancer revealed by next generation sequencing Chang Ya-Sian, Tu Siang-Jyun, Chen Yu-Chia, Liu Ting-Yuan, Lee Ya-Ting, Yen Ju-Chen, Fang Hsin-Yuan, Chang Jan-Gowth. Respiratory Research.2021;22(1). CrossRef
- The prevalence and prognostic value of KRAS co-mutation subtypes in Chinese advanced non-small cell lung cancer patients Cai Dongjing, Hu Chengping, Li Li, Deng Shichao, Yang Jing, Han-Zhang Han, Li Min. Cancer Medicine.2020;9(1). CrossRef
- The prevalence and concurrent pathogenic mutations of K-RAS G12C in Northeast Chinese non-small-cell lung cancer patients. The lung treated with first-line pembrolizumab monotherapy Liu Y, Li H, Zhu J, et al . Cancer Manag Res.2021;13:2447-54.
- Prognostic impact of KRAS mutation status for patients with stage IV adenocarcinoma of the lung treated with first-line pembrolizumab monotherapy Noordhof A. L., Damhuis R. a. M., Hendriks L. E. L., Langen A. J., Timens W., Venmans B. J. W., Geffen W. H.. Lung Cancer (Amsterdam, Netherlands).2021;155. CrossRef
- KRAS-mutation incidence and prognostic value are metastatic site-specific in lung adenocarcinoma: poor prognosis in patients with KRAS mutation and bone metastasis Lohinai Zoltan, Klikovits Thomas, Moldvay Judit, Ostoros Gyula, Raso Erzsebet, Timar Jozsef, Fabian Katalin, Kovalszky Ilona, Kenessey István, Aigner Clemens, Renyi-Vamos Ferenc, Klepetko Walter, Dome Balazs, Hegedus Balazs. Scientific Reports.2017;7. CrossRef
- EGFR, KRAS, BRAF, ALK, and cMET genetic alterations in 1440 Sardinian patients with lung adenocarcinoma Colombino Maria, Paliogiannis Panagiotis, Cossu Antonio, Santeufemia Davide Adriano, Sini Maria Cristina, Casula Milena, Palomba Grazia, Manca Antonella, Pisano Marina, Doneddu Valentina, Palmieri Giuseppe. BMC pulmonary medicine.2019;19(1). CrossRef
- The Presence of Concomitant Mutations Affects the Activity of EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC) Patients Rachiglio Anna Maria, Fenizia Francesca, Piccirillo Maria Carmela, Galetta Domenico, Crinò Lucio, Vincenzi Bruno, Barletta Emiddio, Pinto Carmine, Ferraù Francesco, Lambiase Matilde, Montanino Agnese, Roma Cristin, Ludovini Vienna, Montagna Elisabetta Sara, De Luca Antonella, Rocco Gaetano, Botti Gerardo, Perrone Francesco, Morabito Alessandro, Normanno Nicola. Cancers.2019;11(3). CrossRef
- Actionable Mutation Profiles of Non-Small Cell Lung Cancer patients from Vietnamese population Dang Anh-Thu Huynh, Tran Vu-Uyen, Tran Thanh-Truong, Thi Pham Hong-Anh, Le Dinh-Thong, Nguyen Lam, Nguyen Ngoc-Vu, Thi Nguyen Thai-Hoa, Nguyen Chu Van, Le Ha Thu, Thi Nguyen Mai-Lan, Le Vu Thuong, Nguyen Phuc Huu, Vo Binh Thanh, Thi Dao Hong-Thuy, Nguyen Luan Thanh, Van Nguyen Thien-Chi, Bui Quynh-Tram Nguyen, Nguyen Long Hung, Nguyen Nguyen Huu, Thi Nguyen Quynh-Tho, Le Truong Xuan, Do Thanh-Thuy Thi, Dinh Kiet Truong, Do Han Ngoc, Phan Minh-Duy, Nguyen Hoai-Nghia, Tran Le Son, Giang Hoa. Scientific Reports.2020;10(1). CrossRef
- Frequency and type of epidermal growth factor receptor mutations in moroccan patients with lung adenocarcinoma Errihani Hassan, Inrhaoun Hanane, Boukir Anouar, Kettani Fouad, Gamra Lamia, Mestari Amina, Jabri Lamia, Bensouda Youssef, Mrabti Hind, Elghissassi Ibrahim. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer.2013;8(9). CrossRef
- The Frequency of Epidermal Growth Factor Receptor (EGFR) mutations in Iraqi patients with Non-Small Cell Lung Cancer (NSCLC) Ramadhan Hanan H., Taaban Dhuha F., Hassan Jubran K.. Asian Pacific Journal of Cancer Prevention.2021;22(2). CrossRef
- Epidermal growth factor receptor (EGFR) positive non-small-cell lung carcinoma (NSCLC) patients in the Gulf region: current status, challenges, and call for action Jaafar H, Mohieldin A, Mohsen R, et al . J Cancer Prev Curr Res.2020;11:130-4.
- High epidermal growth factor receptor amplification rate but low mutation frequency in Middle East lung cancer population Al-Kuraya Khawla, Siraj Abdul K., Bavi Prashant, Al-Jommah Naif, Ezzat Adnan, Sheikh Salwa, Amr Samir, Al-Dayel Fouad, Simon Ronald, Guido Sauter. Human Pathology.2006;37(4). CrossRef
- Epidermal growth factor receptor and KRAS mutations in lung adenocarcinoma: a retrospective study of the Lebanese population Fakhruddin Najla, Mahfouz Rami, Farhat Fadi, Tfayli Arafat, Abdelkhalik Rabab, Jabbour Mark, Yehia Lamis, Mahfoud Ziyad, Zaatari Ghazi. Oncology Reports.2014;32(5). CrossRef
- EGFR mutation incidence and characteristics in non-squamous lung carcinoma in the Lebanese population. Kattan Joseph Gergi, Haddad Fady, Kourie Hampig Raphael, Naderi Samah, Rassy Marc, El Karak Fadi Rafic, Ghosn Marwan, Sader-Ghorra Claude. Journal of Clinical Oncology.2015;33(15_suppl). CrossRef
- EGFR mutation status in Middle Eastern patients with non-squamous non-small cell lung carcinoma: A single institution experience Naderi Samah, Ghorra Claude, Haddad Fady, Kourie Hampig Raphael, Rassy Marc, El Karak Fadi, Ghosn Marwan, Abadjian Gérard, Kattan Joseph. Cancer Epidemiology.2015;39(6). CrossRef
- Prevalence of EGFR and ALK Mutations in Lung Adenocarcinomas in the Levant Area - a Prospective Analysis Tfayli Arafat, Rafei Hind, Mina Alain, Khalil Maya, Fakhreddin Najla, Mahfouz Rami, Hamouri Shadi, Farhat Fadi, Salem Ziad, Dbouk Haifa, Rabee Haider, Saghir Nagi, Shamseddine Ali, Makarem Jawad, Bitar Nizar, Mougharbil Anas, Assi Hazem, Temraz Sally, Mukherji Deborah, Matalka Ismail, Zaatari Ghazi. Asian Pacific Journal of Cancer Prevention.2017;18(1). CrossRef
- Comparison of ARMS and direct sequencing for detection of EGFR mutation and prediction of EGFR-TKI efficacy between surgery and biopsy tumor tissues in NSCLC patients Shaozhang Zhou, Ming Zhou, Haiyan Peng, Aiping Zeng, Qitao Yu, Xiangqun Song. Medical Oncology (Northwood, London, England).2014;31(5). CrossRef
- Mode of action and application of Scorpion primers to mutation detection Thelwell N., Millington S., Solinas A., Booth J., Brown T.. Nucleic Acids Research.2000;28(19). CrossRef
- Detection of PCR products using self-probing amplicons and fluorescence Whitcombe D., Theaker J., Guy S. P., Brown T., Little S.. Nature Biotechnology.1999;17(8). CrossRef
- EGFR Mutation Detection and Its Association With Clinicopathological Characters of Lung Cancer Patients Gaur Priyanka, Bhattacharya Sandeep, Kant Surya, Kushwaha R. a. S., Singh Gaurav, Pandey Sarika. World Journal of Oncology.2018;9(5-6). CrossRef
- EGFR mutation status in a series of Turkish non-small cell lung cancer patients Calibasi-Kocal Gizem, Amirfallah Arsalan, Sever Tolga, Umit Unal Olcun, Gurel Duygu, Oztop Ilhan, Ellidokuz Hulya, Basbinar Yasemin. Biomedical Reports.2020;13(2). CrossRef
- Epidermal growth factor receptor mutation (EGFRMUT) in non-small cell lung cancer (NSCLC) in the Middle East Jazieh AR, Jaafar HN, Mustafa R, et al . J Clin Oncol.2013;31:e19035.
- Patterns of epidermal growth factor receptor mutation in non-small-cell lung cancers in the Gulf region Jazieh Abdul Rahman, Jaafar Hasan, Jaloudi Mohammed, Mustafa Rasha Saleh, Rasul Kakil, Zekri Jamal, Bamefleh Hanaa, Gasmelseed Ahmed. Molecular and Clinical Oncology.2015;3(6). CrossRef
- KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes in NSCLC: A meta-analysis of 41 studies Pan Wei, Yang Yan, Zhu Hongcheng, Zhang Youcheng, Zhou Rongping, Sun Xinchen. Oncotarget.2016;7(7). CrossRef
- Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib Jackman David M., Yeap Beow Y., Sequist Lecia V., Lindeman Neal, Holmes Alison J., Joshi Victoria A., Bell Daphne W., Huberman Mark S., Halmos Balazs, Rabin Michael S., Haber Daniel A., Lynch Thomas J., Meyerson Matthew, Johnson Bruce E., Jänne Pasi A.. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2006;12(13). CrossRef
- The different clinical significance of EGFR mutations in exon 19 and 21 in non-small cell lung cancer patients of China Li M., Zhang Q., Liu L., Liu Z., Zhou L., Wang Z., Yue S., Xiong H., Feng L., Lu S.. Neoplasma.2011;58(1). CrossRef
- Relationship between epidermal growth factor receptor gene mutations and clinicopathological features in patients with non-small cell lung cancer in western Turkey Unal Olcun Umit, Oztop Ilhan, Calibasi Gizem, Baskin Yasemin, Koca Dogan, Demir Necla, Akman Tulay, Ellidokuz Hulya, Yilmaz Ahmet Ugur. Asian Pacific journal of cancer prevention: APJCP.2013;14(6). CrossRef
- Overall Survival Analysis and Characterization of an EGFR Mutated Non-Small Cell Lung Cancer (NSCLC) Population Aguiar Filipa, Fernandes Gabriela, Queiroga Henrique, Machado José Carlos, Cirnes Luís, Souto Moura Conceição, Hespanhol Venceslau. Archivos De Bronconeumologia.2018;54(1). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details