Characterization of Multiple Omics Signatures in Relation to Dietary Pattern for in Silico Personalised Colon Cancer Risk Stratification: Study Protocol for a Case-control Study and the Challenges Faced During the COVID-19 Pandemic
Download
Abstract
Background: Personalised nutrition and medicine are the future of healthcare. In relation to cancer, public and healthcare professionals often seek dietary recommendations for cancer prevention. Among the important cancers that can be prevented by diet and lifestyle is colorectal cancer (CRC). CRC is one of the commonest cancers globally, and is a major health concern in Malaysia as it presents with high mortality and morbidity rates, causing a significant socioeconomic burden to the country. While extensive research has been conducted on the treatment and mechanisms of cancer, there have been no reports on the associations between metabolites, novel biomarkers of cancer, and dietary patterns in the context of cancer prevention in the Malaysian multiethnic population.
Methods: A case control study will be conducted in Malaysia, involving patients diagnosed with CRC, colorectal adenoma and a group of healthy participants. Multiple endpoints will be analyzed, namely metabolomic signatures, epigenetic marks, inflammatory markers and relationship with dietary patterns will be established. Multiple machine learning models will then be used to develop personalised risk stratification algorithms. Recruitment began in July 2019 and is ongoing due to COVID-19 pandemic.
Discussion: This study will be the first to identify alterations in metabolites, inflammatory markers and epigenetic marks associated with dietary patterns and CRC risk in Malaysia. Understanding on how dietary patterns influence CRC risk in the multi-ethnic Malaysian population and identification of novel oncometabolites for CRC risk, will allow for development of personalised evidence-based recommendations in reducing individual risks of CRC.
Introduction
Colorectal cancer (CRC) is a global public health concern, particularly in Malaysia as data in Globocan showed that in 2020, the incidence reported is 3816 for colon cancer incidence and 2690 for rectal cancer incidence and projected to increase by 20% for both cancer cases. Sadly, the mortality rate is also projected to increase in numbers by the year 2040 [1]. In addition to dietary habits, other factors such as lifestyle and nutritional status greatly influence the incidence of CRC [2, 3]. CRC is the fifth most common cause of malignant death worldwide after lung, stomach, liver, and breast cancers [4] and late detection is one of the main reasons the survival rate, particularly in Malaysia, is low [5].
CRC is an important public health problem, particularly in low- and middle- income countries. About one in three people diagnosed with CRC die of the disease within 5 years after diagnosis [6]. In addition to the human cost, the tangible and intangible costs of treating CRC are enormous. Improvements in surgical modalities and adjuvant chemotherapy have increased the cure rates in early-stage disease, but a significant portion of the patients will develop recurrence or advanced disease [7]. Although there has been a marked reduction in CRC incidence and death rates across developed countries due to improved screening services and specialized care [6], screening and early detection services for CRC are lacking in Malaysia. Furthermore, apart from a small number of CRC cases related to genetic disposition and advanced age, up to 90% of CRC cases are non-hereditary, and have been linked to modifiable behavioral practices related to diet, physical inactivity, obesity, heavy alcohol consumption and tobacco use [8-10].
Many studies on Western populations have investigated the association between diet and lifestyle factors and CRC [11-13]. Modern approaches are focused on the analysis of dietary patterns, which may better capture the association between diet and cancer risk. This type of approach may deal with the correlation and potential effect modifications of the diet composition, considering the variety of nutrients consumed. A priori dietary patterns are used in hypothesis-driven analyses that evaluate the effects adherence to a predetermined pattern of food consumption is assumed to have on health [14]. However, evidence on colorectal adenomas has only been reviewed and at the very least observational studies have quantified the summary effects of adherence to dietary patterns on this pre-cancerous condition. In a systematic review conducted recently with 12 studies were reviewed, dietary pattern was suggested to have an association with the risk of colorectal adenomas [15].
At the molecular level, genomic instability is key characteristics of most cancer cells, and has been extensively described in the context of cancer and various health related outcomes [16]. There is an increased tendency of genome alteration during cell division. Cancer frequently results from damage of multiple genes controlling cell division and tumor suppressors. The maintenance of genomic stability is essential for cellular integrity to prevent errors from DNA replication, endogenous genotoxic stress such as reactive oxygen species (ROS) from cellular metabolism, and exogenous carcinogen insults; such as ultraviolet light, processed food, ionizing radiation or DNA damaging chemicals. In a genome-wide association study (GWAS) conducted on CRC tissues, genes that were found to attribute to the multiple mechanisms that are responsible for normal and abnormal controls of gene expression, including those related to mutation, promoter methylation and miRNA expression hence leading to genomic instability [17]. Recent cancer prevention report by the International Agency for Research on Cancer (IARC) emphasized on various possible steps that can achieve optimum healthy habit hence reducing genetic damage via holistic approach inclusive of diet, physical activity, body mass index and lifestyle habits [18]. Our previous study on zinc carnosine has shown that antioxidant agents can be used as cancer chemo-preventive alternatives thanks to their anti-inflammatory potential and protective effects against genome integrity as well as their potential to reduce oxidative stress in the cells particularly in colorectal cancer cell lines [19].
In Malaysia, evidence from small scales studies have shown that abdominal obesity is one the significant risk factors for CRC [20, 21]. Meanwhile abdominal obesity and hypercholesterolemia synergically doubled the risk of CRC [21]. When screening for a large number of polymorphism and environmental factors that may contribute to development of CRC in Malaysia, it was found that 23 SNPs were significantly associated with colorectal cancer risk [22]. In the same study, among the dietary risk factors investigated, high intake of red meat was found to be significantly associated with increased risk of CRC.
Although screening and lifestyle play important roles in CRC prevention, identifying a causal mechanism of mutagenesis is essential to understand the complex pathway involved and to develop personalized prevention strategies [23]. The gut microbiota has recently been implicated in CRC pathogenesis and is useful for personalized prevention. In addition, it has been documented that majority of CRC risk factors including diet (high red meat / high fat / low fiber), obesity, physical activity, smoking, and alcohol use also have significant effects on the gut microbial community [24]. Because the gut microbiota alters the metabolic environment of the host, it is predicted that it may indirectly influence genomic instability events, and thus carcinogenesis. Hale and colleagues reported that increased sugar, protein, and lipid metabolism along with increased bile acid production could promote a colonic environment that supports the growth of bile-tolerant microbes that may produce genotoxic or inflammatory metabolites, which could play a role in catalyzing adenoma development and eventually CRC [25].
To our knowledge, no other recent reports have comprehensively detailed the relationship between CRC and lifestyle habits in Malaysia particularly in the context of exploring dietary patterns and other bioindicators in the precancerous group as well as in newly diagnosed CRC patients. Malaysia is characterized by different ethnicity and origins; consequently, different lifestyles, health status, working places and incomes could have divergent effects on the incidence of CRC. Therefore, studying the possible risk factors of the aforementioned variables as well as family history of CRC could shed light on the pathobiology of CRC and thus possible prevention measures.
Cancer epidemiology literature is currently saturated, especially those coming from developed countries. Major questions remain as to whether it is possible to recommend a one-size-fits-all approach to healthy dietary patterns for cancer prevention according to changes in cancer-related biomarkers across genetically and demographically diverse populations. A challenge to studying the biological mechanisms underpinning the effects of these dietary combinations lies in the modelling of dietary patterns that is appropriate to the human diet. The question whether to tailor dietary recommendations to subgroups of the population based on susceptibility factors (family history, sex, age, other lifestyle mediators, metabolomic signatures, epigenetic marks) remains unanswered and will be important in advancing our knowledge of CRC. Hence, this article will focus on the study protocol of in silico risk stratification based on multiple omics signature in relation to dietary pattern for colorectal cancer risk prevention.
Materials and Methods
Design
A multi-center case control study will be conducted and is useful for the study of colorectal cancer as it can provide data on risk factors in a relatively cost-efficient manner. This allow an in-depth interview to be conducted examining the dietary pattern in the past, before the disease developed and other possible variables that may affect the relationship between diet and CRC. Exposures of interest include dietary pattern, anthropometry and physical assessment, physical inactivity and biological markers as presented in Figure 1.
Figure 1. Simplified Diagram of Study Design.
Three patient groups will be enrolled, namely patients diagnosed with colorectal adenoma, newly diagnosed CRC patients and a group of healthy participants.
Sampling Methods
Convenience sampling method
Inclusion and exclusion criteria
Cases will be selected among patients aged 18 years to 79 years old with neoplastic polyps which include adenomas, traditional and sessile serrated polyps who had visited the outpatient oncology clinic or were admitted into the oncology wards within the period of the study in Hospital Canselor Tuanku Muhriz (HCTM), Hospital Kuala Lumpur (HKL) and Hospital Sultanah Nur Zahirah (HSNZ), Terengganu. It will also involve newly diagnosed patients with primary invasive colorectal cancer at any stage of T0-4, N0, M0 according to American Joint Committee on Cancer (AJCC) TNM system as in Table 1.
| AJCC Stage | Stage grouping | Stage description* |
| 0 | Tis | The cancer is in its earliest stage. This stage is also known as carcinoma in situ or intramucosal carcinoma (Tis). It has not grown beyond the inner layer (mucosa) of the colon or rectum. |
| N0 | ||
| M0 | ||
| I | T1 or T2 | The cancer has grown through the muscularis mucosa into the submucosa (T1), and it may also have grown into the muscularis propria (T2). It has not spread to nearby lymph nodes (N0) or to distant sites (M0). |
| N0 | ||
| M0 | ||
| IIA | T3 | The cancer has grown into the outermost layers of the colon or rectum but has not gone through them (T3). It has not reached nearby organs. It has not spread to nearby lymph nodes (N0) or to distant sites (M0). |
| NO | ||
| M0 | ||
| IIB | T4a | The cancer has grown through the wall of the colon or rectum but has not grown into other nearby tissues or organs (T4a). It has not yet spread to nearby lymph nodes (N0) or to distant sites (M0). |
| N0 | ||
| M0 | ||
| IIC | T4b | The cancer has grown through the wall of the colon or rectum and is attached to or has grown into other nearby tissues or organs (T4b). It has not yet spread to nearby lymph nodes (N0) or to distant sites (M0). |
| N0 | ||
| M0 | ||
| IIIA | T1 or T2 | The cancer has grown through the mucosa into the submucosa (T1), and it may also have grown into the muscularis propria (T2). It has spread to 1 to 3 nearby lymph nodes (N1) or into areas of fat near the lymph nodes but not the nodes themselves (N1c). It has not spread to distant sites (M0). |
| N1/N1c | ||
| M0 | ||
| OR | ||
| T1 | The cancer has grown through the mucosa into the submucosa (T1). It has spread to 4 to 6 nearby lymph nodes (N2a). It has not spread to distant sites (M0). | |
| N2a | ||
| M0 | ||
| T3 or T4a | The cancer has grown into the outermost layers of the colon or rectum (T3) or through the visceral peritoneum (T4a) but has not reached nearby organs. It has spread to 1 to 3 nearby lymph nodes (N1a or N1b) or into areas of fat near the lymph nodes but not the nodes themselves (N1c). It has not spread to distant sites (M0). | |
| N1/N1c | ||
| M0 | ||
| OR | ||
| IIIB | T2 or T3 | The cancer has grown into the muscularis propria (T2) or into the outermost layers of the colon or rectum (T3). It has spread to 4 to 6 nearby lymph nodes (N2a). It has not spread to distant sites (M0). |
| N2a | ||
| M0 | ||
| OR | ||
| T1 or T2 | The cancer has grown through the mucosa into the submucosa (T1), and it might also have grown into the muscularis propria (T2). It has spread to 7 or more nearby lymph nodes (N2b). It has not spread to distant sites (M0). | |
| N2b | ||
| M0 | ||
| T4a | The cancer has grown through the wall of the colon or rectum (including the visceral peritoneum) but has not reached nearby organs (T4a). It has spread to 4 to 6 nearby lymph nodes (N2a).It has not spread to distant sites (M0). | |
| N2a | ||
| M0 | ||
| OR | ||
| IIIC | T3 or T4a | The cancer has grown into the outermost layers of the colon or rectum (T3) or through the visceral peritoneum (T4a) but has not reached nearby organs. It has spread to 7 or more nearby lymph nodes (N2b). It has not spread to distant sites (M0). |
| N2b | ||
| M0 | ||
| OR | ||
| T4b | The cancer has grown through the wall of the colon or rectum and is attached to or has grown into other nearby tissues or organs (T4b). It has spread to at least one nearby lymph node or into areas of fat near the lymph nodes (N1 or N2). It has not spread to distant sites (M0). | |
| N1 or N2 | ||
| M0 | ||
| IVA | Any T | The cancer may or may not have grown through the wall of the colon or rectum (Any T). It might or might not have spread to nearby lymph nodes. (Any N). It has spread to 1 distant organ (such as the liver or lung) or distant set of lymph nodes, but not to distant parts of the peritoneum (the lining of the abdominal cavity) (M1a). |
| Any N | ||
| M1a | ||
| IVB | Any T | The cancer might or might not have grown through the wall of the colon or rectum (Any T). It might or might not have spread to nearby lymph nodes (Any N). It has spread to more than 1 distant organ (such as the liver or lung) or distant set of lymph nodes,but not to distant parts of the peritoneum (the lining of the abdominal cavity) (M1b). |
| Any N | ||
| M1b | ||
| IVC | Any T | The cancer might or might not have grown through the wall of the colon or rectum (Any T). It might or might not have spread to nearby lymph nodes (Any N). It has spread to distant parts of the peritoneum (the lining of the abdominal cavity), and may or may not have spread to distant organs or lymph nodes (M1c). |
| Any N | ||
| M1c |
* The following additional categories are not listed in the table above; • TX, Main tumor cannot be assessed due to lack of information. • T0, No evidence of a primary tumor. • NX, Regional lymph nodes cannot be assessed due to lack of information.
Patient recruitment will be based on the classification made by experts in the field, namely gastroenterologists and surgeons involved in each hospital.
Inclusion criteria: Control
1. Age 18 - 79 years old
2. Not on a special diet regime
3. Able to communicate and carry out the interview
4. Not having any chronic diseases
Case 1
1. Age 18 - 79 years old
2. Patient diagnosed with neoplastic polyps which include adenomas, traditional and sessile serrated polyps
3. Not on a special diet regime
4. Able to communicate and carry out the interview
Case 2
1. Age 18 - 79 years old
2. Newly diagnosed colorectal cancer patient with primary invasive colorectal cancer at any stage except for advanced or metastatic cancer (N1-3, M1)
3. Not on a special diet regime
4. Able to communicate and carry out the interview
Exclusion criteria
Exclusion included those diagnosed with other types of cancer (except colon and rectal).
Sample Size Calculation
Allowing an error rate of 2.5%, a level of significance (type 1 error) of 5% and 95% confidence interval and with an a priori estimate of 10% prevalence of CRC in controls and a least odds ratio of 2.35 [26], the OpenEpi sample size calculator indicated that a sample size of 291 (97 participants in each control group, case 1 group and case 2 group) is required to achieve a strong power of 80 % in this study. Taking into consideration the 30% drop outs, we estimate that a sample size of 130 volunteers in each group would be sufficient thus a total of 390 participants for this study.
Data Collection
Self-administered or interview-based questionnaires will be administered to respondents. Questionnaires include socio-demographic variables, information on diet (Diet History Questionnaire with multiple checklists of dietary carcinogen and pro inflammatory diet) and physical activity (International Physical Activity Questionnaire- Short Form). Estimated duration for answering all questionnaires is 30 – 45 minutes.
Anthropometric measurements including body weight, height, waist circumference and body composition will be measured using standard methods and trained researchers. Specific body composition measures will be measured using Inbody Bio analyser.
For novel blood biomarkers, we will collect 10 ml non-fasted blood. Blood plasma will be used for metabolomic analysis and inflammatory markers, while plasma and buffy coat samples will be used for epigenetic marks (global methylation).
Dietary Pattern Analysis Dietary Data
A validated version of dietary history questionnaire (DHQ) [27] will be used in this study to determine dietary patterns. Subjects will be interviewed by a dietitian to gather detailed information about their dietary habits including the type, amount, method of cooking and frequency. This includes all foods and beverages taken in their daily routines, including snacks or beverages taken between major meals. In addition, the subjects are also asked about the frequency of having each mealtime, ranging from every day, almost every day (5 to 6 times per week), sometime (3 to 4 times per week), seldom (once to twice per week), very seldom (less than once a week) and never. The time, venue and with whom the meal will be taken and recorded. During diet history assessment, respondents will also be probed using dietary carcinogen / ultra-processed food group and pro inflammatory dietchecklist as in Table 2.
| Pro- inflammatory food | Food that may contain carcinogen/Ultra-processed | |
| Red meat | Sweet / savoury packaged snacks | Noodles / rice vermicelli / kuey teow / laksa |
| Processed meat | Ice cream | Fishballs / meatballs/ vegetable balls |
| Organ meat | Chocolate | Instant noodles |
| Fish, non-fatty | Sweets / Chewing gum | Jelly / pudding |
| Egg | Carbonated drinks | Pizza |
| Sweet drink | Packed bread / bun | Sausage / hotdogs |
| White rice | Margerine / spread | Burger |
| Bread | Biscuits | Nuggets |
| Noodles | Cake and pastry | Chicken / meat stock cubes / instant sauce |
| Margerine | Breakfast cereals | Weight loss / Health supplements / drinks |
| French fries | Energy bar / energy drink | Alcoholic beverages |
| Flavoured milk (Pack) | (Whiskey, gin rum, vodka) | |
| Fruits Yoghurt | ||
| Cocoa drinks | ||
| Fruit cordial / drinks |
Information gathered from DHQ is analysed by using Nutritionist Pro (Axxya Systems Stafford, USA) software. In addition, to detect under-reporting, energy intake was divided by the basal metabolic rate (energy intake / basal metabolic rate).
Physical activity
It is well established that physical activity is one of the factors that convincingly decrease the risk of colorectal cancer as reported by World Cancer Research Fund and American Institute for Cancer Research [18]. In addition, physical activity is also reported to be one of the important factors to improve the prognosis of CRC [28]. In this study, a validated short last 7 days self- administered version of the International Physical Activity Questionnaires (IPAQ) will be used to obtain information on physical activity level.
Multiple Omics Platform Analysis
1. Metabolomics analysis
a. Untargeted Profiling of Serum Samples by High- Resolution Mass Spectrometry
The samples will be processed in random order by mixing 20 µL of serum with 200 µL of acetonitrile; filtered with 0.2 µm Agilent Captiva ND filter plate and analyzed by high-resolution LC–MS as previously described. In short, sample extracts will be injected (2 µL) into a UHPLC–MS system consisting of a 1290 Binary LC system, a Jet Stream ESI source, and a 6550 QTOF mass spectrometer (Agilent Technologies, Santa Clara, CA). A Waters Acquity UPLC HSS T3 reverse phase column (2.1 × 100 mm, 1.8 µm) will be used for chromatographic separation by a water: methanol gradient over a 13-min run. The mass spectrometer will be operated in positive or negative ionization mode and the data were acquired over a mass range of 50 to 1000 Da. Conditions for positive ionization mode will be as follows: drying gas (nitrogen) temperature 175 °C and flow 12 L min–1, sheath gas temperature 350 °C and flow 11 L min–1, nebulizer pressure 45 psi, capillary voltage 3500 V, nozzle voltage 300 V, and fragmentor voltage 175V. Acquisition rate will be set at 1.67 Hz. Conditions for negative ionization mode will be the same except for negative polarity. MS/MS spectra will be acquired using 1.3 Da isolation width with 10, 20, and 40 V collision energies. A quality control (QC) serum pool will be prepared by mixing small aliquots of all study samples; QC samples prepared from the pool will be processed along with the study samples. Blank samples and QC samples will be incorporated into the analytical run at regular intervals to monitor method performance. QC samples will be used to assess overall technical variability through calculation of coefficients of variability for a small subset of known features for all QC samples in the batch. All samples will be injected twice in two separate batches with the mass spectrometer operating in positive and negative ionization modes, respectively.
b. Preprocessing of Mass Spectrometry Data
Molecular features will be extracted from positive and negative ionization mode datasets using Agilent recursive feature extraction workflow as described earlier. In brief, molecular feature extraction algorithms for small molecules will be employed with peak height threshold 800 and 600 counts for positive and negative mode data, respectively, with ions limited to [M+H]+ and [M– H]−. Alignment will be performed with retention time and mass windows of 0.07 min and 15 ppm ± 2 mDa, respectively. Targets for the recursive extraction will be created from features with chromatographic peak heights ≥15 000 and
≥2000 counts in at least 2% of samples for positive and negative mode data, respectively. For recursive analysis, features will be re-extracted with ±10 ppm and ±0.05 min matching tolerance without intensity threshold. Mass will be calculated from ions >10% of peak height and <10% above saturation and final alignment will be performed using the same parameters as above.
c. Identification of Metabolites
Retained features will be first grouped by chromatographic retention time and intensity correlation across subjects, since multiple features often originate from the same metabolite. This will result in several clusters of related masses, which were then annotated cluster by cluster. Mass differences between cluster members will be calculated to search for known fragments and adducts. The m/z values were searched using 8 ppm tolerance in online metabolite databases such as the Human Metabolome Database (HMDB; http:// www.hmdb.ca), the Metlin metabolite database, and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Annotations will be confirmed by matching of retention time and MS/MS spectra with those of authentic chemical standards wherever possible. The resulting chromatograms were inspected, manually reintegrated where needed, and the resulting peak areas were used as a measurement of intensity for statistical analyses.
2. Inflammatory markers
Plasma samples obtained from the participants will be aliquoted and stored at 80C for analysis. According to their role as inflammatory biomarkers, circulating factors such as insulin-like growth factor-1 (IGF-1), soluble tumor necrosis factor receptor 2 (sTNFR-2), and interleukin-8 (IL-8) are proposed to mediate the inflammatory response linked to CRC within the tumor microenvironment. Based on the manufacturer’s instructions, all inflammatory markers will be measured using Enzyme-Linked Immunosorbent Assay (ELISA) kits as follows: IGF-1 (Catalog No: ab211651) from Abcam (Cambridge, UK), sTNFR-2 (Catalog No: ELH-TNFR2) from RayBiotech (Norcross, GA, USA), and IL-8 (Catalog No: ELH-IL8) from RayBiotech.
3. Global DNA Methylation
Genomic DNA will be isolated from blood biospecimens using the QIAamp DNA Mini and blood kit (Qiagen Inc., USA) in accordance with the manufacturer’s instructions. DNA quantity and purity will be determined by spectrophotometric absorbance. Global genomic DNA methylation levels will be determined using the 5-mC DNA ELISA kit (Zymo Research, USA) in accordance with the manufacturer’s instructions. The assay relies on a sensitive and specific anti-5-methylcytosine monoclonal antibody which would allow for the accurate quantitation of 5-methylcytosine in DNA samples without the need for bisulfite conversion. Briefly, 100 ng of DNA will be prepared for each assay reaction. DNA will be denatured and applied to the wells of the ELISA plate in the presence of a coating buffer. Samples will be incubated with an anti-5-methylcytosine primary antibody and a horseradish peroxidase conjugate secondary antibody, with the relevant washing steps. Following the addition of a HRP substrate, the quantity of 5-methylcytosine in each sample will be determined based on its absorbance measured at 405 nm using an ELISA plate reader against a series of standards which will be generated using mixtures of the negative and positive control samples included with the kit. Each biospecimen will be assayed in duplicate.
In silico risk stratification - Multiple machine learning model
Several machine learning algorithms will be developed, namely neural network (NN), random forest (RF), support vector machine (SVM), neuro-fuzzy (NF), and logistic regression models. The performance of each model will be assessed by cross-validation on test sets of known sample groupings. Following the testing stage, the best method could be applied to new patients, which is expected to return the risk probability of the disease or the exact prognosis of the class (case/control) label. The outcome of the analysis was CRC diagnosis.
Statistical Analysis
The student unpaired t-test and correlation coefficients will be used for mean comparisons. The odds ratios (OR) and 95% confidence intervals (CI) obtained from multivariable logistic regression models will be taken as the measures of predictors of CRC. All statistical analyses will be conducted using IBM SPSS for Windows version 25.0 and statistical significance will be set at p-value of ≤ 0.05.
Challenges during COVID19 pandemic
To date, data collection for this study has been hampered due to COVID19 pandemic. In March 2020, as the COVID-19 pandemic hit Malaysia, universities closed their doors with no information about when they would reopen. A Movement Control Order was implemented by the government. While many undergraduate students
learning, most researchers were asked to carry on with their work from home, and research progress plateaud. Most researchers are affected during this crisis, and this includes data collection in hospitals. Hospital Canselor Tuanku Muhriz UKM, which is the main hospital for data collection, was designated for COVID19 cases and was closed to any research. Patient movements were restricted and data collection for this study was halted for almost one year. This year (2021), cases are on the rise again and another series of Movement Control Order was deployed hence affecting our data collection (Figure 2).
Figure 2. Recruitment of Subjects.
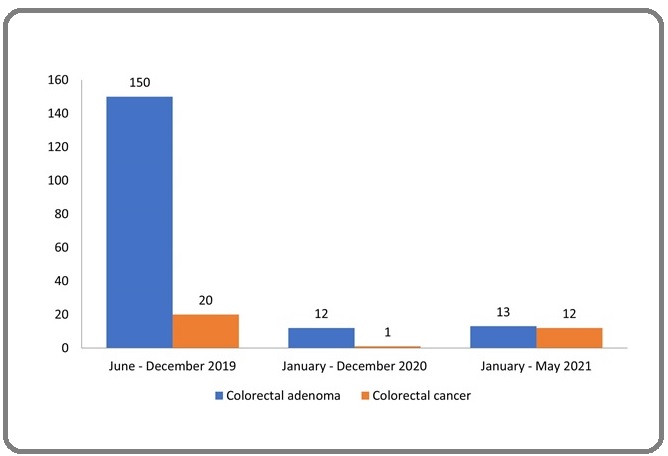
Patients are not keen to be interviewed via phone call and video as our patient demographics encompass those of older age.
Patient and public involvement
When first designing the study, the research team received input from patient representatives who had experienced colorectal cancer in addition to professionals in National Cancer Research Institute. All parties had input into the original grant proposal and played a part in developing and agreeing the research questions, study procedures (eg, inclusion criteria) and choice of primary outcome.
Patients are involved during data collection and at the end of data collection, and they will be called for a group meeting to discuss interpretation of the study. We will also discuss plan of translating the data and data dissemination with the involved stakeholders.
Discussion
In Malaysia, colorectal cancer is one of the most frequently diagnosed cancers and a leading cause of death with a low rate of survivorship. The current screening strategy in Malaysia include the immunochemical Faecal Occult Blood Test (iFOBT) and colonoscopy. However, the effectiveness of these screening programme was hindered by some barriers such as clients’ reluctance to undergo screening due to uncomfortableness of procedure, personal privacy and hygiene related to the collection of faecal specimens [29, 30]. Therefore, predicting colon cancer risk is vital as a mitigating strategy. To date, this is the first study which supports a holistic and comprehensive study design for colorectal cancer risk. The strength of this study is on the outcome as all the data will provide a platform for developing personalised risk stratification using modifiable lifestyle factors. This study will thoroughly assess the effects of various risk factors of CRC which include dietary factor, physical activity and body fatness. Identification and profiling of biomarkers such as genome stability, inflammatory molecules and metabolomics as well as on the epigenetics will provide the evidence related to the aforementioned risk factors of CRC. Understanding this association will help in evidence-based guidance on how to improve prevention of CRC incidents. Awareness for CRC screening for better prognosis and early detection is still poor aware in Malaysia and a complete and organized CRC screening guideline for the country is tremendously needed [31]. The outcome from this study will generate the necessary information to develop a complete CRC screening guideline and therefore will greatly reduce the economic burden of CRC. Evidence-based recommendations are crucially needed to empower patient to lead a better lifestyle, to improve their prognosis, reduce CRC risk and for a better health outcome. In conclusion, the results of this study will be the initial step in what might become a bigger multicenter international research programme enrolling people with life-threatening cancer diagnoses from various populations.
Acknowledgments
We acknowledge the contributions of the funding body and all cooperation’s given at the study site involving patients, nurses and all individuals involved in this study who wish to remain anonymous.
Abbreviations
CRC, Colorectal cancer; SNPs, Single nucleotide polymorphisms
Funding
The study was supported by Internal Research University Grant (UKM GUP 2018-065), Ministry of Higher Education (FRGS/1/2020/STG02/UKM/02/5) and UICC Technical Fellowship 2019.
Author contributions
RS was responsible for overall conceptualization, initial draft and revising draft for content. NMAM, SNB, RARA, SS, NFR, FC, ZAMA, ANAK and MRS were responsible for conceptualization and revising draft for content. All the authors have read and approved the final manuscript.
Competing interests
The authors report no conflict of interest related to the work.
Consent for publication
Not applicable
Ethics approval and consent to participate
This study received approval from the Medical Research and Ethics Committees (MREC), PPUKM (JEP-2019-243) and (National Medical Research Register ethical approval (NMRR-19-4208-50423). Informed consent will be obtained from each patient prior to enrolment.
References
- GLOBOCAN. Cancer Tomorrow: Institute Agency for Research on Cancer; 2020 [Available from: https://gco.iarc.fr/tomorrow/en/dataviz/trendstypes=0_1&sexes=1_2&mode=cancer&group_populations=1&multiple_populations=1&multiple_cancers=1&cancers=8&populations=458&age_start=3..
- Nutrients, foods, and colorectal cancer prevention Song Mingyang, Garrett Wendy S., Chan Andrew T.. Gastroenterology.2015;148(6). CrossRef
- Trends in colorectal cancer incidence and related lifestyle risk factors in 15-49-year-olds in Canada, 1969-2010 Patel Parth, De Prithwish. Cancer Epidemiology.2016;42. CrossRef
- Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle, Siegel Rebecca L., Torre Lindsey A., Jemal Ahmedin. CA: a cancer journal for clinicians.2018;68(6). CrossRef
- Malaysian Study on Cancer Survival (MyScan) MYSCAN . National Cancer Registry, National Cancer Institute, Ministry of Health Malaysia; 2018..
- Cancer screening in the United States, 2016: A review of current American Cancer Society guidelines and current issues in cancer screening Smith Robert A., Andrews Kimberly, Brooks Durado, DeSantis Carol E., Fedewa Stacey A., Lortet-Tieulent Joannie, Manassaram-Baptiste Deana, Brawley Otis W., Wender Richard C.. CA: a cancer journal for clinicians.2016;66(2). CrossRef
- Pharmacogenomics DNA Biomarkers in Colorectal Cancer: Current Update Ab Mutalib Nurul-Syakima, Md Yusof Najwa F., Abdul Shafina-Nadiawati, Jamal Rahman. Frontiers in Pharmacology.2017;8. CrossRef
- Association of adherence to lifestyle recommendations and risk of colorectal cancer: a prospective Danish cohort study Kirkegaard Helene, Johnsen Nina Føns, Christensen Jane, Frederiksen Kirsten, Overvad Kim, Tjønneland Anne. BMJ (Clinical research ed.).2010;341. CrossRef
- Multiple health behaviours: overview and implications Spring Bonnie, Moller Arlen C., Coons Michael J.. Journal of Public Health (Oxford, England).2012;34 Suppl 1. CrossRef
- A DASH dietary pattern and the risk of colorectal cancer in Canadian adults Jones-McLean E., Hu J., Greene-Finestone L. S., Groh M.. Health Promotion and Chronic Disease Prevention in Canada: Research, Policy and Practice.2015;35(1). CrossRef
- Lifestyle factors associated with survival after colorectal cancer diagnosis Boyle T., Fritschi L., Platell C., Heyworth J.. British Journal of Cancer.2013;109(3). CrossRef
- Meta-analyses of colorectal cancer risk factors Johnson Constance M., Wei Caimiao, Ensor Joe E., Smolenski Derek J., Amos Christopher I., Levin Bernard, Berry Donald A.. Cancer causes & control: CCC.2013;24(6). CrossRef
- Impact of lifestyle factors and nutrients intake on occurrence of gastrointestinal cancer in Tunisian population Baroudi Olfa, Chaaben Arij Ben, Mezlini Amel, Moussa Amel, Omrane Ines, Jilson Irene, Benammar-Elgaaied Amel, Chabchoub Soufia. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine.2014;35(6). CrossRef
- Dietary patterns and breast cancer: a review with focus on methodological issues Edefonti Valeria, Randi Giorgia, La Vecchia Carlo, Ferraroni Monica, Decarli Adriano. Nutrition Reviews.2009;67(6). CrossRef
- Dietary patterns and risk of colorectal adenoma: a systematic review and meta-analysis of observational studies Godos J, Bella F, Torrisi A, Sciacca S, Galvano F, Grosso G. Journal of human nutrition and dietetics : the official journal of the British Dietetic Association.2016;29(6). CrossRef
- DNA Damage, Copper and Lead Associates with Cognitive Function among Older Adults Meramat A., Rajab N. F., Shahar S., Sharif R. A.. The Journal of Nutrition, Health & Aging.2017;21(5). CrossRef
- Integrated Analysis of Copy Number Variation and Genome-Wide Expression Profiling in Colorectal Cancer Tissues Hassan Nur Zarina Ali, Mokhtar Norfilza Mohd, Sin Teow Kok, Rose Isa Mohamed, Sagap Ismail, Harun Roslan, Jamal Rahman. PLOS ONE.2014;9(4). CrossRef
- World Cancer Research Fund, American Institute for Cancer Research. Diet, nutrition, physical activity and colorectal cancer London: World Cancer Research Fund/ American Institute for Cancer Research; 2018 [109] Available from: https://www.wcrf.org/diet-and-cancer/..
- Antioxidant, Anti-inflammatory, and Genomic Stability Enhancement Effects of Zinc l-carnosine: A Potential Cancer Chemopreventive Agent? Ooi Theng Choon, Chan Kok Meng, Sharif Razinah. Nutrition and Cancer.2017;69(2). CrossRef
- Food intake and colorectal adenomas: a case-control study in Malaysia Ramadas Amutha, Kandiah Mirnalini. Asian Pacific journal of cancer prevention: APJCP.2009;10(5).
- A case-control study on the association of abdominal obesity and hypercholesterolemia with the risk of colorectal cancer Ulaganathan Vaidehi, Kandiah Mirnalini, Shariff Zalilah Mohd. Journal of Carcinogenesis.2018;17. CrossRef
- Role of genetic & environment risk factors in the aetiology of colorectal cancer in Malaysia Ramzi Nurul Hanis, Chahil Jagdish Kaur, Lye Say Hean, Munretnam Khamsigan, Sahadevappa Kavitha Itagi, Velapasamy Sharmila, Hashim Nikman Adli Nor, Cheah Soon Keat, Lim Gerard Chin Chye, Hussein Heselynn, Haron Mohd Roslan, Alex Livy, Ler Lian Wee. The Indian Journal of Medical Research.2014;139(6).
- 16S rRNA Gene Sequencing for Deciphering the Colorectal Cancer Gut Microbiome: Current Protocols and Workflows Osman Muhammad-Afiq, Neoh Hui-Min, Ab Mutalib Nurul-Syakima, Chin Siok-Fong, Jamal Rahman. Frontiers in Microbiology.2018;9. CrossRef
- Gut microbiome and colorectal adenomas Dulal Santosh, Keku Temitope O.. Cancer Journal (Sudbury, Mass.).2014;20(3). CrossRef
- Shifts in the Fecal Microbiota Associated with Adenomatous Polyps Hale Vanessa L., Chen Jun, Johnson Stephen, Harrington Sean C., Yab Tracy C., Smyrk Thomas C., Nelson Heidi, Boardman Lisa A., Druliner Brooke R., Levin Theodore R., Rex Douglas K., Ahnen Dennis J., Lance Peter, Ahlquist David A., Chia Nicholas. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology.2017;26(1). CrossRef
- Dietary patterns and colorectal cancer risk in a Korean population: A case-control study Park Yoon, Lee Jeonghee, Oh Jae Hwan, Shin Aesun, Kim Jeongseon. Medicine.2016;95(25). CrossRef
- Validation of a Dietary History Questionnaire against a 7-D Weighed Record for Estimating Nutrient Intake among Rural Elderly Malays Shahar S., Earland J., Abdulrahman S.. Malaysian Journal of Nutrition.2000;6(1).
- Nutrition and physical activity guidelines for cancer survivors Rock Cheryl L., Doyle Colleen, Demark-Wahnefried Wendy, Meyerhardt Jeffrey, Courneya Kerry S., Schwartz Anna L., Bandera Elisa V., Hamilton Kathryn K., Grant Barbara, McCullough Marji, Byers Tim, Gansler Ted. CA: a cancer journal for clinicians.2012;62(4). CrossRef
- Overview of colorectal cancer screening programme in Malaysia Arunah C., Feisul I. M., Nor Saleha I. T., Muhammad Radzi A. H.. The Medical Journal of Malaysia.2020;75(3).
- Perceived Deterrence Towards Colonoscopy for Colorectal Cancer Screening among Northern Malaysia Population: A Qualitative Study Mohd Suan Mohd Azri, Tan Wei Leong, Ismail Ibtisam, Abu Hassan Muhammad Radzi. Asian Pacific journal of cancer prevention: APJCP.2020;21(5). CrossRef
- Colorectal cancer in Malaysia: Its burden and implications for a multiethnic country Veettil Sajesh K., Lim Kean Ghee, Chaiyakunapruk Nathorn, Ching Siew Mooi, Abu Hassan Muhammad Radzi. Asian Journal of Surgery.2017;40(6). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details