Screening and Identification of Key Genes in Hepatitis B Virus-Related Hepatocellular Carcinoma Through an Integrated Bioinformatics Approach
Download
Abstract
Objective: Primary liver cancer is one of the main causes of cancer mortality globally, with hepatocellular carcinoma (HCC) being the most frequent type. Chronic hepatitis B virus (HBV) infection is leading cause of HCC. This study aimed to identify significant genes for predicting prognosis in HBV-associated HCC.
Methods: The GSE121248 gene expression profile was obtained from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) for HBV-associated HCC were identified by analyzing this expression profile. Enrichment analyses were performed to discover the role of DEGs in biological processes, cell components, molecular functions, and pathways. Then, protein-protein interaction (PPI) was constructed and 5 hub genes were identified. Finally, survival analysis was conducted to validate the prognostic value of these genes.
Results: A total of 20188 official gene symbols were found, and 119 DEGs were identified between HBV-associated HCC and normal liver tissues. The PPI network identified CCNB1, CDK1, TOP2A, RACGAP1, and ASPM as hub genes. Kaplan-Meier curves showed that the high expression of the hub genes had significantly lower survival.
Conclusion: CCNB1, CDK1, TOP2A, RACGAP1, and ASPM could be potential prognostic biomarkers and therapeutic targets for HBV-associated HCC.
Introduction
Primary liver cancer is one of the main causes of cancer mortality globally [1], with hepatocellular carcinoma (HCC) being the most frequent type [2]. Chronic liver problems such as chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, alcohol-related liver disease, nonalcoholic fatty liver disease (NAFLD), and cirrhosis are leading triggers of HCC [3]. Over the last few decades, significant efforts have been made to establish and improve HCC prevention and monitoring for at-risk groups. Neonatal HBV vaccination has been shown to lower HBsAg seroprevalence in HBV-endemic locations and is currently recommended in the majority of nations [4, 5]. HCC surveillance in patients with cirrhosis using ultrasonography has been suggested by numerous liver disease societies [6, 7]. Currently, hepatectomy and liver transplantation are the primary surgical treatments for HCC patients, in addition, radiation or chemotherapy are frequently employed as adjuvant or palliative treatment [8]. However, conventional medical methods do not suit all individuals with HCC. As a result, a better knowledge of the underlying processes of pathophysiology and tumorigenesis for this disease at the molecular level might aid in the development of innovative early diagnostic and therapeutic therapies to enhance the management and outcome of HCC patients.
Gene profiling and signatures, which can swiftly find differentially expressed genes (DEGs), have substantially expedited cancer research during the last few decades. Several research have looked at the prognostic implications of array-based genes from HCC, however, few have found gene signatures that indicate poor prognosis for HBV-associated HCC. Thus, the aim of this study was to identify significant genes for predicting prognosis in HBV-associated HCC utilizing publicly available data and integrated bioinformatics tools.
Materials and Methods
Data collection
For this research, we chose Gene Expression Omnibus (GEO), a publicly available collection of gene/microarray profiles. This public database has been used to conduct bioinformatics research on many types of cancers, such as breast cancers [9, 10], lung cancers [11, 12], gastrointestinal cancers [13, 14], and bone cancers [15, 16].
The GSE121248 gene expression profile was obtained from the GEO database repository. This dataset used the platform GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array for the mRNA expression profiling. A total of 70 HCC samples and 37 non-tumor tissue samples were collected from chronic HBV-associated HCC and their adjacent normal tissues in the dataset [17].
Identification of differentially expressed genes (DEGs)
First, the dataset was annotated and standardized by quantiles. Then, the limma package of the R software was used to screen the DEGs. The threshold criterion of DEGs was |log FC|>2 and p<0.05. The Principal Components Analysis (PCA) was visualized by using the FactoMineR and factoextra packages. The M-versus-A (MA) plot, volcano plot, and heatmap of DEGs were plotted using the ggpubr and pheatmap packages.
Enrichment analysis
Functional analysis of the DEGs was conducted to further investigate the biological processes and signal pathways in which the DEGs may be engaged. Using the clusterProfiler package, Gene Ontology (GO) analysis was performed to demonstrate the dominating function of the DEGs from molecular function and biological process. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to determine the relationship between DEGs and signal pathways. In this enrichment analysis, all of the findings were visualized, and a statistically significant difference was defined as P<0.05.
Protein-protein interaction (PPI) and module analysis
The STRING platform was used to conduct PPI network analysis for differential genes [18]. The PPI of the DEGs was built using the STRING platform and analyzed using the Cytoscape program. Cytoscape is an open-source bioinformatics application for visualizing gene and protein molecular interaction networks. Moreover, the Cytoscape plug-in Cytohubba was used to identify the hub genes that are strongly associated with HBV-associated HCC [19].
Survival Analysis
The hub genes’ prognostic value was evaluated using the GEPIA database. GEPIA, an interactive web server, can perform interactive and configurable tasks such as differential expression analysis, profile charting, correlation analysis, patient survival analysis, related gene recognition, and dimensionality reduction analysis [20].
Results
Identification of DEGs
A total of 20188 official gene symbols were found, and each gene’s expression was determined. 119 DEGs were identified between HBV-associated HCC and normal liver tissues using the established criteria, comprising 30 up-regulated genes and 89 down-regulated genes. Figure 1 depicts the volcano map of all genes.
Figure 1. Volcano Plot of the Differentially Expressed Genes (DEGs). Significantly up-regulated genes, significantly down-regulated genes and non-DEGs are displayed as red, blue, and yellow dots, respectively, in HBV-associated HCC and their adjacent normal tissues. The top ten upregulated and downregulated DEGs are highlighted.
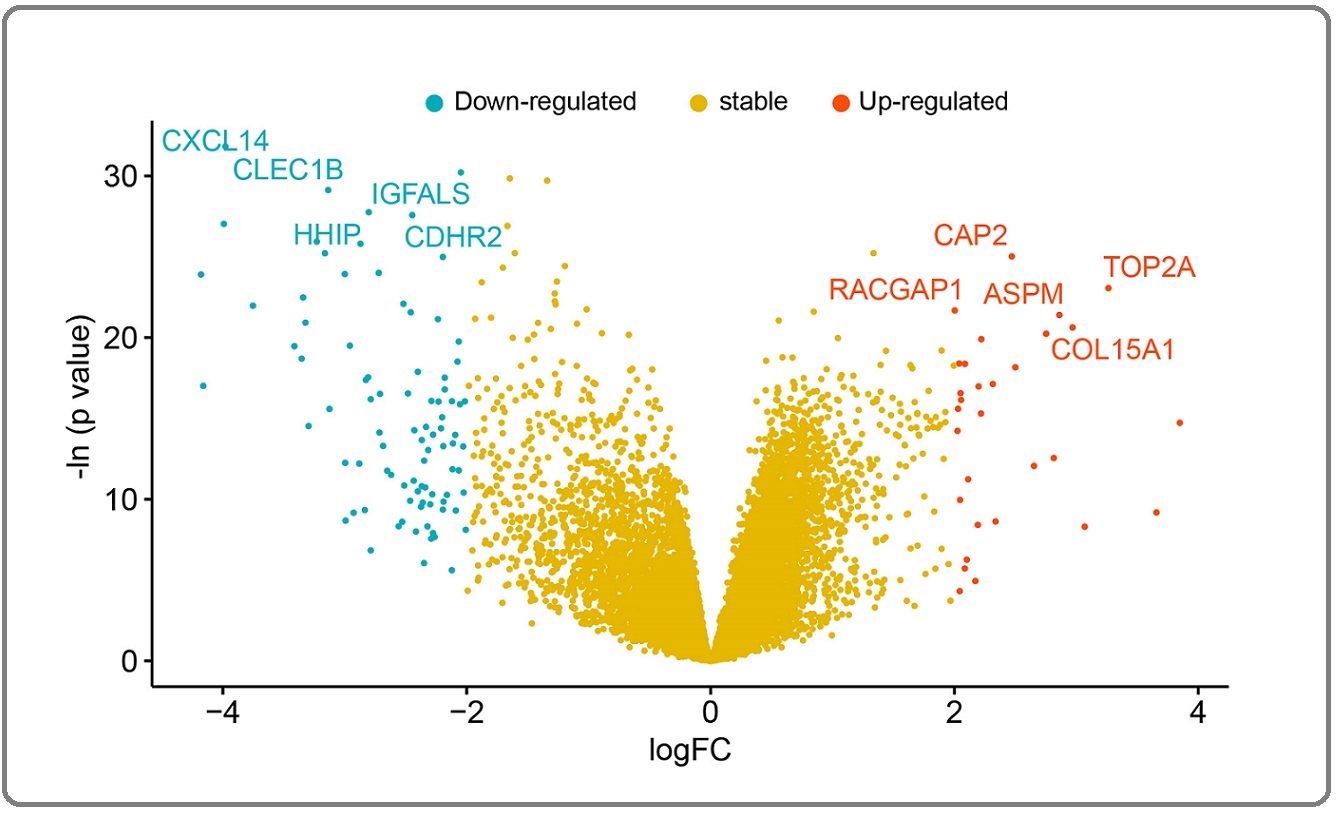
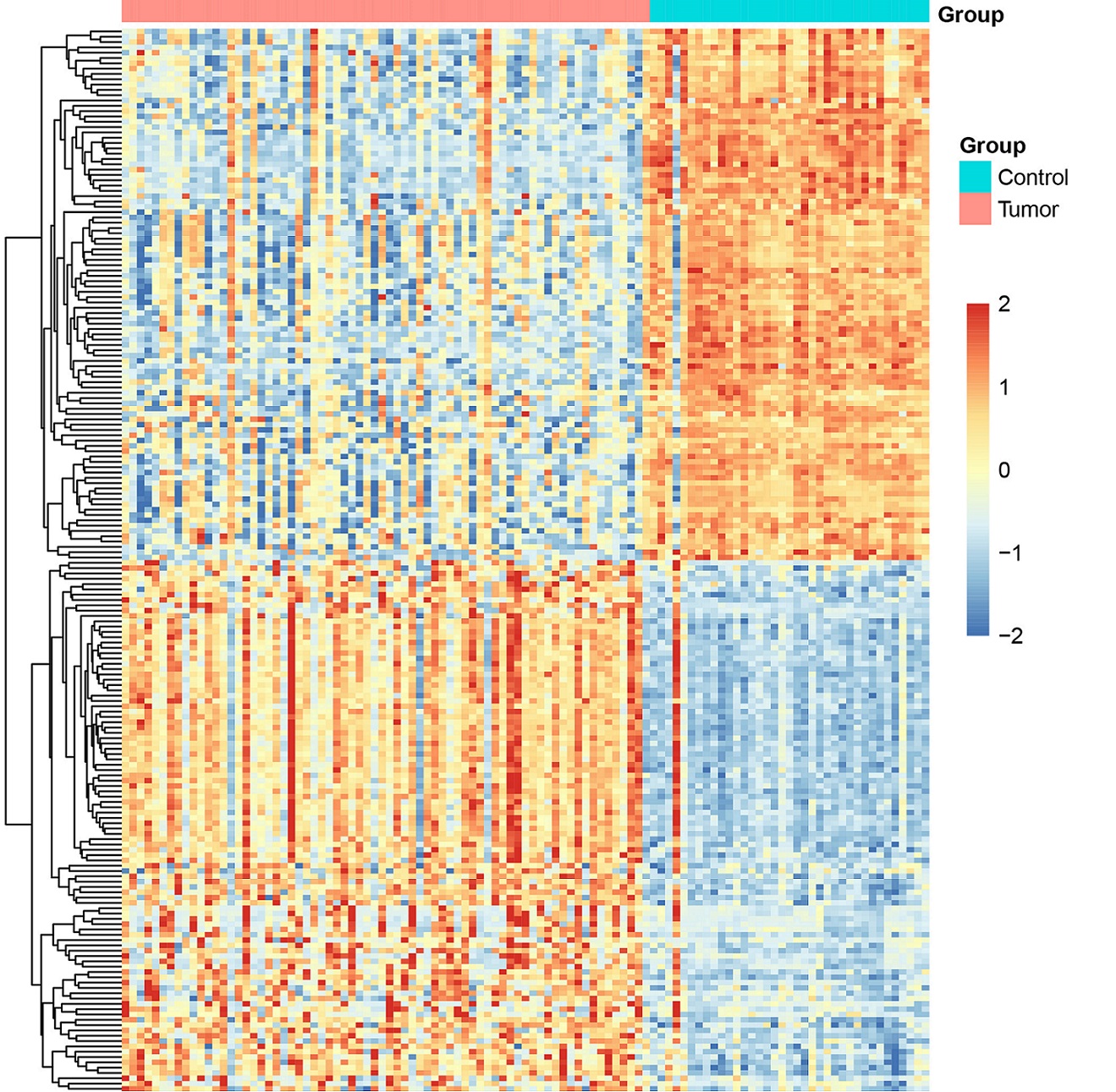
The top 5 up-regulated genes are CAP2, TOP2A, RACGAP1, ASPM and COL15A1, the top 5 down-regulated genes are CXCL14, IGFALS, CLEC1B, HHIP and CDHR2. Figure 2 shows the cluster heatmap of the DEGs.
Figure 2. Cluster Heatmap of the Differentially Expressed Genes (DEGs).
GO and KEGG pathway enrichment analysis
To do GO functional annotation and KEGG pathway enrichment analysis, we utilized the clusterProfiler R package. Figure 3 illustrates the findings of the substantial enrichment analysis. GO analysis results showed that changes in the biological process of DEGs were significantly enriched in regulation of hormone levels, response to xenobiotic stimulus, steroid metabolic process, and cellular hormone metabolic process (Figure 3A).
Figure 3. Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of Differentially Expressed Genes (DEGs). A, Biological Process analysis; B, Cellular Component analysis; C, Molecular Function analysis; D, KEGG pathway analysis.
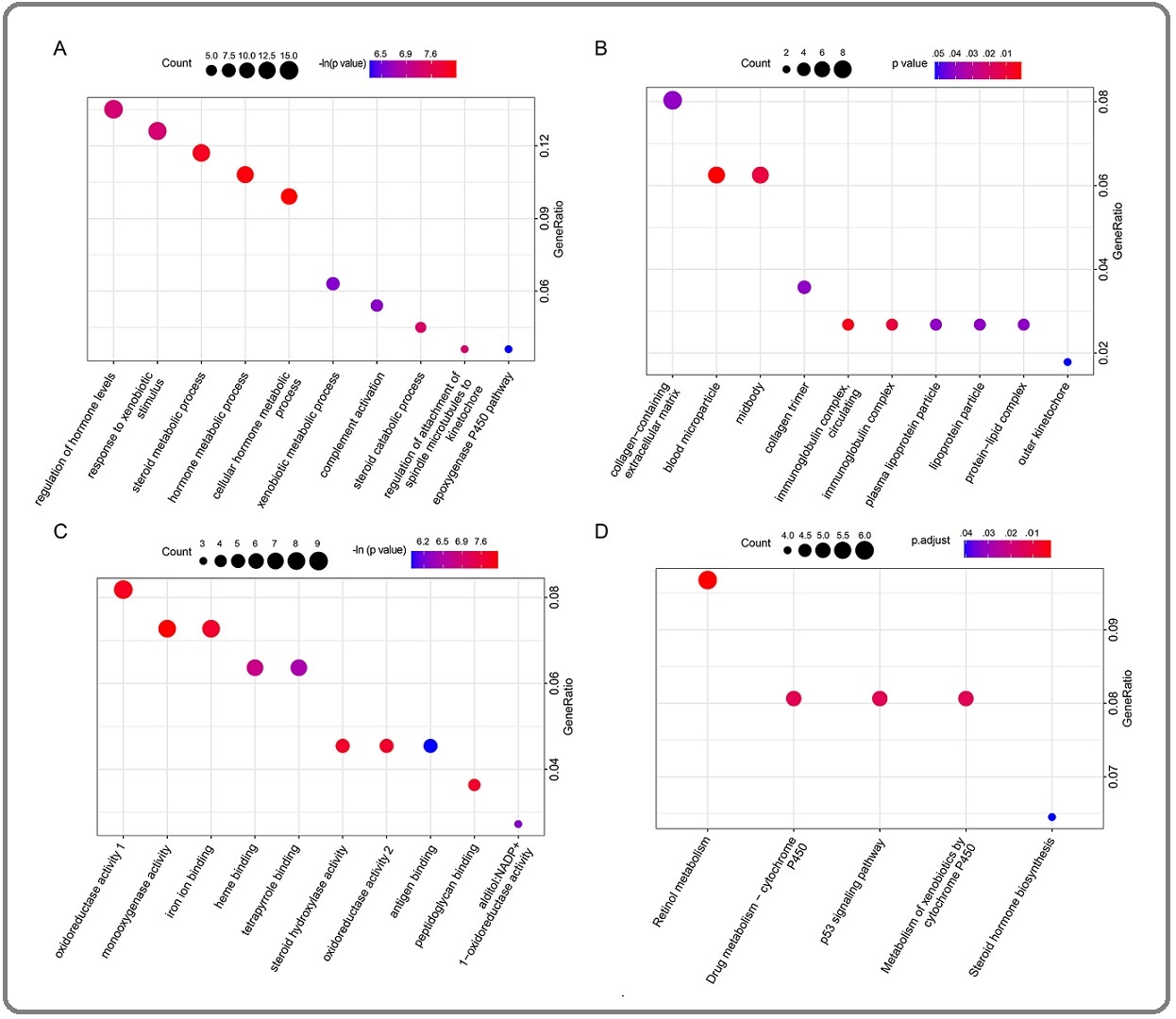
Changes in cell component were mainly enriched in collagen-containing extracellular matrix, blood microparticle, midbody, collagen trimer, and immunoglobulin complex (Figure 3B). Figure 3C showed the changes in molecular function of DEGs, which were mainly enriched in oxidoreductase activity, monooxygenase activity, iron ion binding, heme binding, and tetrapyrrole binding. The results of KEGG pathway analysis (Figure 3D) revealed that DEGs were mostly enriched in retinol metabolism, drug metabolism - cytochrome P450, p53 signaling pathway, metabolism of xenobiotics by cytochrome P450, and steroid hormone biosynthesis.
Construction of PPI Network and identifying of hub genes
The STRING database generated a PPI network with 79 nodes and 212 edges. The PPI was then displayed in Cytoscape to investigate the functional relationships between DEGs (Figure 4A).
Figure 4. Network of Protein-Protein Interaction (PPI). A. PPI network of all DEGs; B. Top 5 hub genes identi- fied using the cytoHubba plug-in.
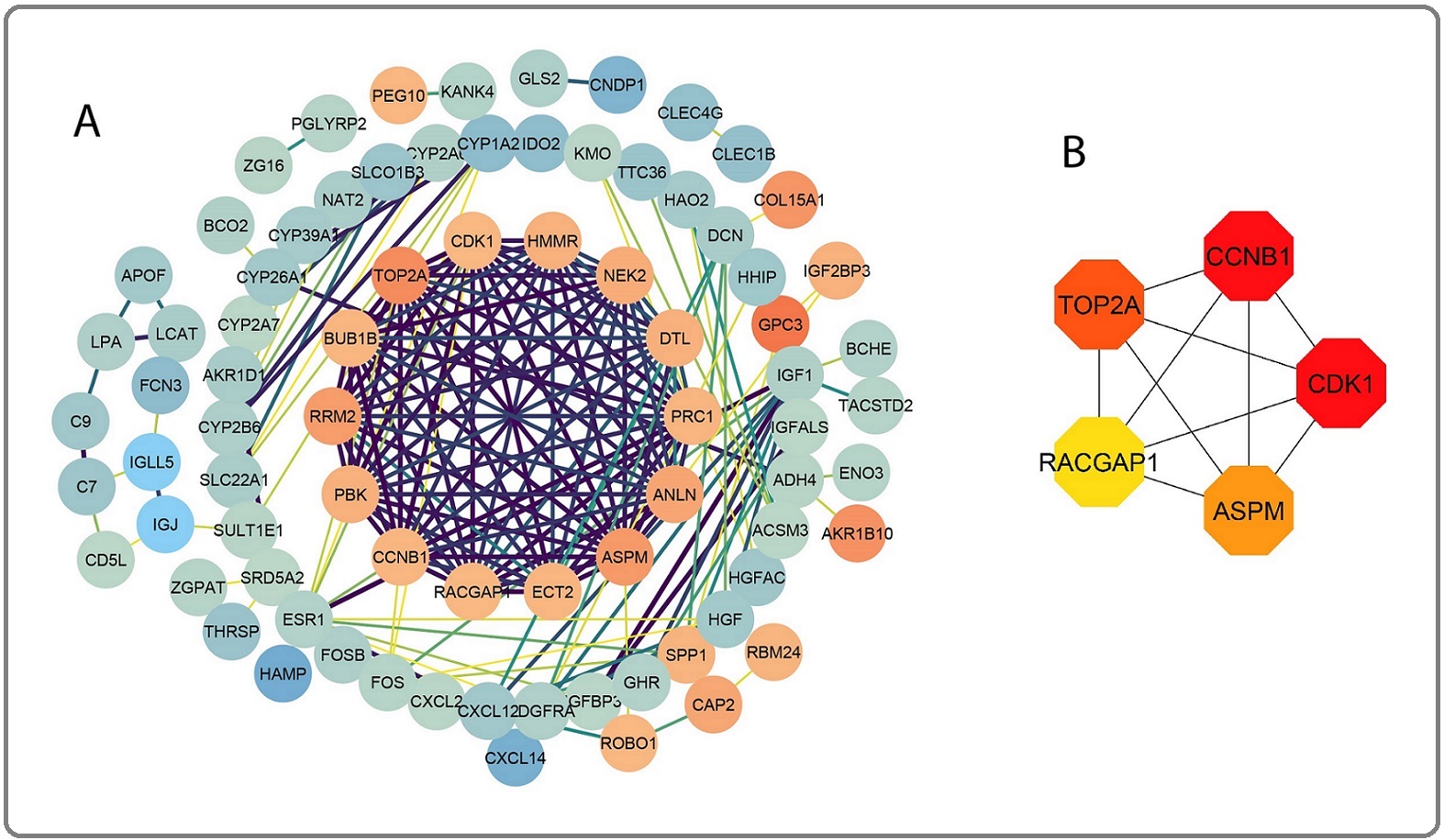
Following that, as demonstrated in Figure 4B, the top five hub genes found in the Cytohubba plug-in using the Maximal Clique Centrality algorithm were CCNB1, CDK1, TOP2A, RACGAP1, and ASPM.
Validation of prognostic value of the hub genes
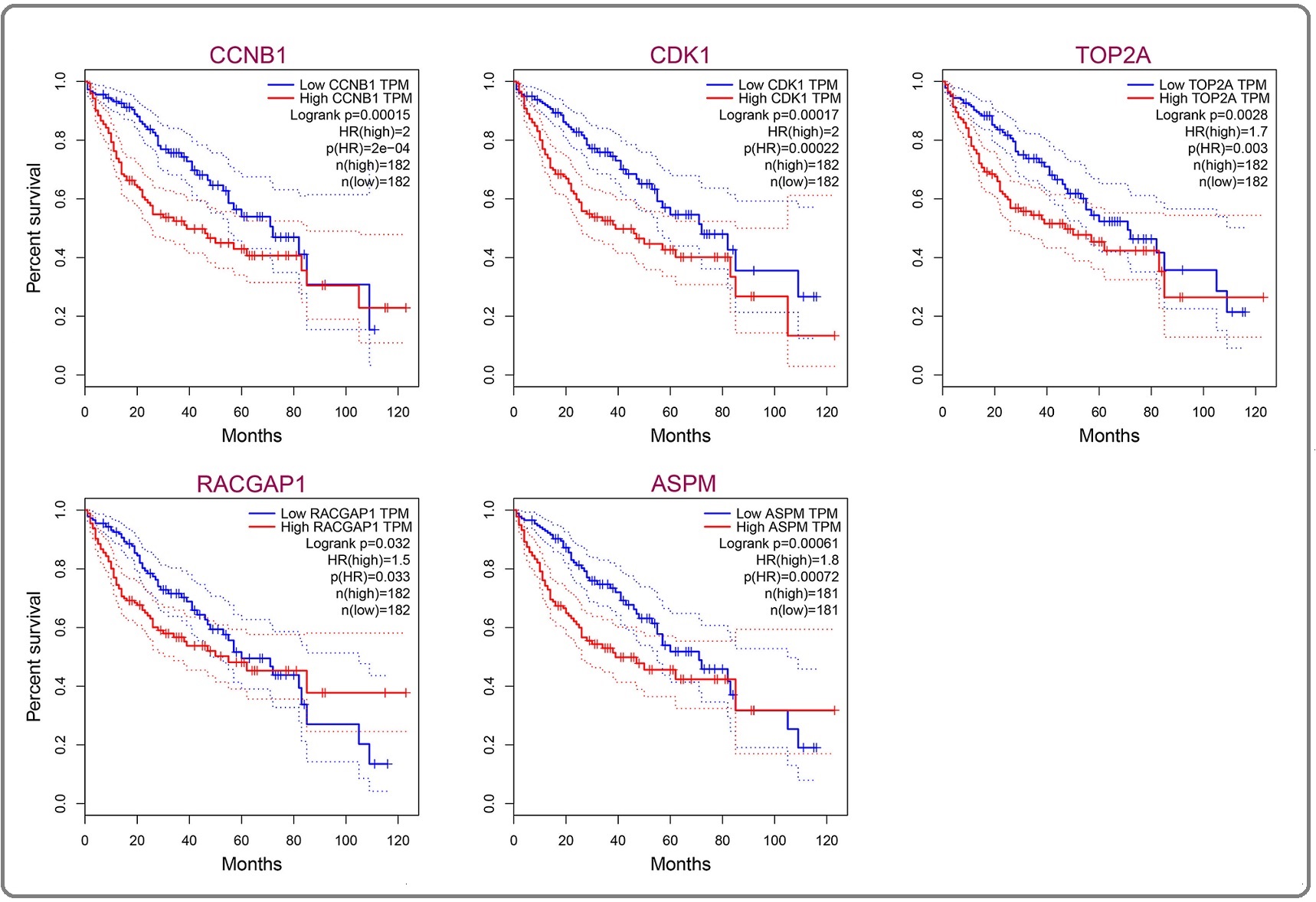
The GEPIA online tool was used to assess the predictive significance of the identified hub genes specific to HBV-associated HCC patients. Based on the medium value of each gene, the patients were divided into the high expression group and low expression group. The Kaplan-Meier curve was used to depict the overall survival analysis between groups. Figure 5 shows that the high expression group of each gene had significantly lower survival than the low expression group (all P<0.05).
Figure 5. Kaplan-Meier Analysis for Overall Survival between High Expression Group and Low Expression Group in Hepatocellular Carcinoma.
Discussion
Despite significant advances in clinical management and pathophysiology prediction for HCC in past few years, the high rate of HCC-specific death remains a significant problem [21]. Chronic HBV infection serves as the most common cause of HCC. In endemic locations, particularly in underdeveloped nations, HBV infection is mostly transmitted vertically. As a result, the typical age of HBV carriers developing HCC is younger than for other etiologies. Furthermore, the majority of patients acquire liver fibrosis and cirrhosis as a result of immunological responses during HBV infection, which promotes the development of HCC. We hoped to get new insights into the molecular mechanisms underpinning HBV-associated HCC formation and progression by using bioinformatics analyses.
This research discovered 119 DEGs including 30 up-regulated genes and 89 down-regulated genes, these DEGs are involved in retinol metabolism, drug metabolism - cytochrome P450, p53 signaling pathway, xenobiotic metabolism via cytochrome P450, and steroid hormone biosynthesis. Furthermore, five hub genes (CCNB1, CDK1, TOP2A, RACGAP1, and ASPM) were verified and found to be related to poorer outcomes in HCC populations with HBV infection.
CCNB1 encoding cyclin B1, a regulatory protein that is important in the mitosis process, is a key member of the conserved cyclin B family [22]. The abnormal activity of this gene can disorganize the cell cycle and cell proliferation, resulting in triggering oncogenesis and cancer development [23, 24]. CCNB1 was shown that substantially related to development, proliferation, migration, and invasion in HCC tumors [25]. Peng et al. [26] indicated that the miR-16/cyclin-B1 axis is the target of zingiberene in the regulation of the growth and invasion of liver tumor cells. Another study also suggests that cyclin B1 may possibly become a biomarker and therapeutic target for HBV-associated HCC [27].
CDK1 is a serine-threonine protein kinase encoding cyclin dependent kinase 1. The abnormal expression of cyclin dependent kinase 1 strongly correlates with carcinogenesis due to its important role in cell mitosis. The antimalarial medicine dihydroartemisinin suppresses the growth of liver cancer cells by lowering CDK1 and CCNB1 expression levels [28]. Wu et al. [29] demonstrated that anti-CDK1 therapy can improve sorafenib anti-tumor response in HCC patient-derived xenograft tumor models by inhibiting CDK1/PDK1/β- Catenin signaling.
TOP2A encodes DNA topoisomerase II alpha, an enzyme that regulates and modifies the topologic states of DNA during transcription. This gene is a target for various anticancer drugs, and a number of mutations in this gene have been linked to the development of treatment resistance [30-32]. TOP2A expression level was shown to be higher in HBV-associated HCC tissues and resulting in a worse prognosis. The findings were consistent with earlier research. Furthermore, the expression of TOP2A in HCC tumors seemed to be associated with the presence of serum HbsAg [33], this might explain why TOP2A was one of the hub genes in HBV-associated HCC.
RACGAP1 encodes a GTPase-activating protein that is part of the centralspindlin complex that regulates cytokinesis, cell proliferation, and differentiation. RACGAP1 has been found as a prognostic biomarker for early HCC identification as well as a therapeutic target for liver cancer and liver cancer stem cells [34]. Wang et al. [35] discovered that the pseudogene RACGAP1P acts as a ceRNA to promote the RACGAP1/Rho/ERK signaling axis, leading to early relapse in HCC. Especially, in patients with HBV-associated HCC, high RACGAP1 expression was linked with an increased risk of mortality [36].
ASPM is the human ortholog of the Drosophila melanogaster abnormal spindle gene, which is required for proper mitotic spindle activity in embryonic neuroblasts. It has been shown to play a role in tumor development. The overexpression of the gene was proven to be a genetic marker indicating HCC’s increased invasive/metastatic potential, a greater likelihood of early tumor recurrence independent of p53 mutation status or tumor stage, and hence a poor prognosis [37]. The N6-methyladenosine modification of ASPM mRNA mediated by METTL3 promoted its expression in liver HCC [38]. ASPM also promotes tumor growth by antagonizing autophagy- mediated Dvl2 degradation and boosting Wnt/-catenin signaling [39].
This study had several following limitations. First, we only used a gene expression profile from the GEO database; this dataset just covers mRNA expression and does not include microRNA, lncRNA, or circRNA expression. This maybe narrows the identification of other potential biomarkers in HBV HCC. Furthermore, additional experiments, both in vivo and in vitro, are required to demonstrate the predictive usefulness of these hub genes.
In conclusion, in the present research, 119 DEGs were found in HBV-associated HCC. Five hub genes, CCNB1, CDK1, TOP2A, RACGAP1, and ASPM, have been identified as potential prognostic biomarkers and therapeutic targets for HBV-associated HCC.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for- profit sectors. The authors declare no conflict of interest.
References
- Hepatocellular Carcinoma Villanueva Augusto. The New England Journal of Medicine.2019;380(15). CrossRef
- Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle, Siegel Rebecca L., Torre Lindsey A., Jemal Ahmedin. CA: a cancer journal for clinicians.2018;68(6). CrossRef
- A global view of hepatocellular carcinoma: trends, risk, prevention and management Yang Ju Dong, Hainaut Pierre, Gores Gregory J., Amadou Amina, Plymoth Amelie, Roberts Lewis R.. Nature Reviews. Gastroenterology & Hepatology.2019;16(10). CrossRef
- Incidence of hepatitis B virus infection in young Chinese blood donors born after mandatory implementation of neonatal hepatitis B vaccination nationwide Tang X., Allain J.-P., Wang H., Rong X., Chen J., Huang K., Xu R., Wang M., Huang J., Liao Q., Shan Z., Luo S., Li T., Li C., Fu Y.. Journal of Viral Hepatitis.2018;25(9). CrossRef
- Long-term impact of infant immunization on hepatitis B prevalence: a systematic review and meta-analysis Whitford Kate, Liu Bette, Micallef Joanne, Yin J. Kevin, Macartney Kristine, Van Damme Pierre, Kaldor John M.. Bulletin of the World Health Organization.2018;96(7). CrossRef
- Critical appraisal of Chinese 2017 guideline on the management of hepatocellular carcinoma Xie Di-Yang, Ren Zheng-Gang, Zhou Jian, Fan Jia, Gao Qiang. Hepatobiliary Surgery and Nutrition.2017;6(6). CrossRef
- Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Hepatology (Baltimore, Md.).2018;68(2). CrossRef
- Association between chemotherapy and prognostic factors of survival in hepatocellular carcinoma: a SEER population-based cohort study Liu M, Liu M, Tang T. Scientific reports.2021;11(1). CrossRef
- A glycolysis-related gene expression signature in predicting recurrence of breast cancer Tang Jianing, Luo Yongwen, Wu Gaosong. Aging (Albany NY).2020;12(24). CrossRef
- Comprehensive Analysis of the Expression and Prognosis for DCTPP1 gene in Breast Cancer Hoang Tien Manh, Bui Thi Thu Hoai, Nguyen Thi Thanh. Asian Pacific Journal of Cancer Biology.2021;6(3). CrossRef
- Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data Yin LI, Jie GU, Fengkai XU, Qiaoliang ZHU, Di GE, Chunlai LU. Scientific Reports.2018;8(1). CrossRef
- Differential Expression and Bioinformatics Analysis of circRNA in Non-small Cell Lung Cancer Sun Qiuwen, Li Xia, Xu Muchen, Zhang Li, Zuo Haiwei, Xin Yong, Zhang Longzhen, Gong Ping. Frontiers in Genetics.2020;11. CrossRef
- Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis Chen L, Lu D, Sun K, Sun Y, Hu P, Li X, Xu F. Gene.2019;692. CrossRef
- Identification of prognosis-related genes in the tumor microenvironment of stomach adenocarcinoma by TCGA and GEO datasets Ren Na, Liang Bin, Li Yunhui. Bioscience Reports.2020;40(10). CrossRef
- The Key Gene Expression Patterns and Prognostic Factors in Malignant Transformation from Enchondroma to Chondrosarcoma Wu Junqing, Huang Yue, Yu Chengxuan, Li Xia, Wang Limengmeng, Hong Jundong, Lin Daochao, Han Xiaoping, Guo Guoji, Hu Tianye, Huang He. Frontiers in Oncology.2021;11. CrossRef
- Hoang TM, Nguyen MT, Chen WS, Zhuang CY, Wang ZX, Wang HQ, et al. Elevated expression of SKP2 correlates with poor prognosis in osteosarcoma: a bioinformatics analysis. Biomed Res Ther, 2021; 8(12), 4782-92. .
- Identification and Validation of a Novel Gene Signature Associated with the Recurrence of Human Hepatocellular Carcinoma Wang Suk, Ooi London, Hui Kam. Clinical cancer research : an official journal of the American Association for Cancer Research.2007;13. CrossRef
- STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene Snel B, Lehmann G, Bork P, Huynen MA. Nucleic acids research.2000;28(18). CrossRef
- cytoHubba: identifying hub objects and sub-networks from complex interactome Chia-Hao CHIN, Shu-Hwa CHEN, Hsin-Hung WU, Chin-Wen HO, Ming-Tat KO, Chung-Yen LIN. BMC Systems Biology.2014;8(4). CrossRef
- GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses Tang Zefang, Chenwei LI, Kang Boxi, Gao Ge, Cheng LI, Zhang Zemin. Nucleic Acids Research.2017;45(W1). CrossRef
- Lymphoid-specific helicase promotes the growth and invasion of hepatocellular carcinoma by transcriptional regulation of centromere protein F expression Yang Xuan, Miao Bi-Si, Wei Chuan-Yuan, Dong Rui-Zhao, Gao Ping-Ting, Zhang Xin-Yu, Lu Jia-Cheng, Gao Chao, Wang Xiao-Ying, Sun Hui-Chuan, Zhou Jian, Fan Jia, Ai-Wu KE, Guo-Ming SHI, Jia-Bin CAI. Cancer Science.2019;110(7). CrossRef
- Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor Suzuki Takashi, Urano Tomohiro, Miki Yasuhiro, Moriya Takuya, Akahira Jun-ichi, Ishida Takanori, Horie Kuniko, Inoue Satoshi, Sasano Hironobu. Cancer Science.2007;98(5). CrossRef
- CREPT/RPRD1B associates with Aurora B to regulate Cyclin B1 expression for accelerating the G2/M transition in gastric cancer Ding Lidan, Yang Liu, Yuqi HE, Bingtao ZHU, Fangli REN, Xuanzi FAN, Wang Yinyin, Mengdi LI, Jun LI, Kuang Yanshen, Sihan LIU, Zhai Wanli, Danhui MA, Yanfang JU, Quentin LIU, Baoqing JIA, Sheng Jianqiu, Chang Zhijie. Cell Death & Disease.2018;9(12). CrossRef
- Cyclin B1 acts as a tumor microenvironment-related cancer promoter and prognostic biomarker in hepatocellular carcinoma Hou Yangming, Wang Xin, Wang Junwei, Sun Xuemei, Liu Xinbo, Hu Han, Fan Wenzhe, Zhang Xinchen, Wu Dequan. The Journal of International Medical Research.2021;49(5). CrossRef
- MicroRNA-144 inhibits cell proliferation, migration and invasion in human hepatocellular carcinoma by targeting CCNB1 Junsheng GU, Xiaorui LIU, Juan LI, Yuting HE. Cancer Cell International.2019;19. CrossRef
- Zingiberene targets the miR-16/cyclin-B1 axis to regulate the growth, migration and invasion of human liver cancer cells Peng Xiulan, Luo Renfeng, Li Jingtao, He Anbing, Wang Xia, Wan Huan, Cai Yahong, Dong Weiguo, Lin Jun. Journal of B.U.ON.: official journal of the Balkan Union of Oncology.2020;25(4). CrossRef
- Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection Weng Li, Du Juan, Zhou Qinghui, Cheng Binbin, Li Jun, Zhang Denghai, Ling Changquan. Molecular Cancer.2012;11. CrossRef
- Anti-malarial drug dihydroartemisinin downregulates the expression levels of CDK1 and CCNB1 in liver cancer Hao Liyuan, Li Shenghao, Peng Qing, Guo Yinglin, Ji Jingmin, Zhang Zhiqin, Xue Yu, Liu Yiwei, Shi Xinli. Oncology Letters.2021;22(3). CrossRef
- Blocking CDK1/PDK1/β-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma Wu Chuan Xing, Wang Xiao Qi, Chok Siu Ho, Man Kwan, Tsang Simon Hing Yin, Chan Albert Chi Yan, Ma Ka Wing, Xia Wei, Cheung Tan To. Theranostics.2018;8(14). CrossRef
- TOP2A and CENPF are synergistic master regulators activated in cervical cancer Beiwei YU, Chen Long, Zhang Weina, Yue LI, Zhang Yibiao, Gao Yuan, Teng Xianlin, Wang Qian, Jia Hongtao, Liu Xiangtao, Zheng Hui, Hou Ping, Yu Hongyan, Sun Ying, Zhang Zhiqin, Zhang Ping, Zhang Liqin. BMC Medical Genomics.2020;13(1). CrossRef
- Mutual regulation of MDM4 and TOP2A in cancer cell proliferation Liu Tao, Zhang Hailong, Yi Sha, Lubing GU, Zhou Muxiang. Molecular Oncology.2019;13(5). CrossRef
- TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup Labbé DP, Sweeney CJ, Brown M, Galbo P, Rosario S, Wadosky KM, Sjöström M, et al . Clinical cancer research : an official journal of the American Association for Cancer Research.2017;23(22). CrossRef
- TOP2A amplification and overexpression in hepatocellular carcinoma tissues Panvichian Ravat, Tantiwetrueangdet Anchalee, Angkathunyakul Napat, Leelaudomlipi Surasak. BioMed Research International.2015;2015. CrossRef
- PRC1 and RACGAP1 are Diagnostic Biomarkers of Early HCC and PRC1 Drives Self-Renewal of Liver Cancer Stem Cells Liao Shixin, Wang Kaili, Zhang Lulu, Shi Gaoli, Wang Zhiwei, Chen Zhenzhen, Zhu Pingping, He Qiankun. Frontiers in Cell and Developmental Biology.2022;10.
- Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence Wang Meng-Yao, Chen Dong-Ping, Qi Bin, Li Ming-Yi, Zhu Yan-Yi, Yin Wen-Jing, He Lu, Yu Yi, Li Zhou-Yu, Lin Ling, Yang Fang, Lin Zhi-Rui, Liu Jin-Quan. Cell Death & Disease.2019;10(6). CrossRef
- Identification of genes predicting unfavorable prognosis in hepatitis B virus-associated hepatocellular carcinoma Sha Meng, Cao Jie, Zong Zhi-Peng, Xu Ning, Zhang Jian-Jun, Tong Ying, Xia Qiang. Annals of Translational Medicine.2021;9(12). CrossRef
- ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma Lin Shih-Yeh, Pan Hung-Wei, Liu Shu-Hsiang, Jeng Yung-Ming, Hu Fu-Chang, Peng Shian-Yang, Lai Po-Lin, Hsu Hey-Chi. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research.2008;14(15). CrossRef
- METTL3‐mediated m6A methylation of ASPM drives hepatocellular carcinoma cells growth and metastasis Wang An, Chen Xiaofeng, Li Dongen, Yang Liang, Jiang Jianshuai. Journal of Clinical Laboratory Analysis.2021;35(9). CrossRef
- ASPM promotes hepatocellular carcinoma progression by activating Wnt/β-catenin signaling through antagonizing autophagy-mediated Dvl2 degradation Zhang H, Yang X, Zhu L, Li Z, Zuo P, Wang P, et al . FEBS open bio.2021;11(10). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details