Philadelphia Negative MPNs, MDS/MPNs and MDS in the Context of Recent WHO Changes and Inclusion of Molecular Signatures into Prognostic Tools: A Single Centre Experience
Download
Abstract
Introduction: The fifth edition of WHO classification of myeloid neoplasms has introduced major changes in the defining criteria and grouping of MDS, MDS/MPNs and MPNs. Recently published literature has also cited the importance of new risk-scoring systems by integrating genomic profiling with hematologic and cytogenetic characteristics, in order to improve the prognostic discrimination of patients and represents a valuable tool for clinical decision-making.
Aim: To find out the prevalence and molecular spectrum of Philadelphia-negative MPNs, MDS/MPNs and MDS in our subset of patients and henceforth to evaluate the impact on diagnosis, risk stratification and treatment decision-making of patients.
Methods: This retrospective observational study included all newly diagnosed patients of non-Philadelphia positive MPNs, MDS, and MPN/MDS, in whom complete baseline diagnostic work-up was available including complete blood counts, bone marrow morphology and biopsy, cytogenetic and molecular studies.
Results: The most frequent entities in our cohort of patients were primary myelofibrosis (32.8%), MDS (32.8%) and CMML (16.4%). In PIMF, 50% patients were JAK2- mutated while 30% were triple negative (JAK-, CALR-, MPL-). The commoner epigenetic modifiers among MPNs were ASXL1, TET2 and IDH2. The predominant CMML molecular signatures in our patients were NRAS, U2AF2, SETBP1, ASXL and SH2B3. There was no significant effect of WHO changes and recently introduced molecular scoring models on the diagnosis and risk stratification of all these MPN and MDS/MPN patients However, in MDS and PIMF patients, recent WHO subtyping plus IPSS-M & GIPSS scoring respectively enabled refining of risk groups.
Conclusion: Molecular profiling helps in better risk stratification of patients across all groups as well as in making therapeutic decisions. However, in resource constrained settings, it is not always possible to stratify patients on the basis of molecular signatures and hence, scoring models such as DIPSS and IPSS-R holds their ground strongly even today for offering appropriate therapy to patients without compromising on quality care.
Introduction
The fifth edition of WHO classification of myeloid neoplasms has introduced major changes in the defining criteria and grouping of myelodysplastic syndromes (MDS), myelodysplastic/ myeloproliferative neoplasms (MDS/MPNs) and myeloproliferative neoplasms (MPNs) [1]. Moreover, recently published literature has also cited the importance of the mutation combined with revised International Prognostic Scoring System (MIPSS-R) scoring in MDS over IPSS-R by integrating genomic profiling with hematologic and cytogenetic characteristics, to improve the prognostic discrimination of patients and represents a valuable tool for clinical decision-making. It incorporates additional mutations such as IDH2, RUNX1, NRAS, ETV6, U2AF1, SRSF2, DNMT3A, ASXL1, SF3B1, CBL, NPM1, FLT3 etc which contribute to the clonal heterogeneity of MDS patients [2,3]. Similarly, new prognostic models such as genetically inspired prognostic scoring system (GIPSS) have been implemented, incorporating adverse karyotypic abnormalities such as -7, inv3, i (17q), +21, +19, 12p-, 11q- as well as high molecular risk mutations such as ASXL1, SRSF2, EZH2, IDH and U2AF1 in comparison to the previous dynamic IPSS (DIPSS) scoring model. GIPSS is based exclusively on mutations and karyotype [4].
In the background of all these changes, we tried to find out the prevalence and molecular spectrum of Philadelphia-negative MPNs, MDS/MPNs and MDS in our subset of patients and henceforth to evaluate the impact of the new changes on diagnosis, risk stratification and treatment decision-making of patients. There is very limited data on this topic and especially from this part of the world.
Materials and Methods
This retrospective observational study included all newly diagnosed patients of non-Philadelphia positive MPNs, MDS, and MPN/MDS who presented in the Hemato-Oncology OPD during the period between July 2020-June 2022, in whom complete baseline diagnostic work-up was available including complete blood counts, bone marrow morphology and biopsy, cytogenetic and molecular studies and flow cytometry wherever applicable. Ethical clearance was obtained for the conduct of the study. Cytogenetics was done by direct/overnight harvesting of bone marrow aspirate, GTG banding and chromosomal analysis using the karyotype software (Applied Spectral Imaging, India).
For Next Generation Sequencing assay, DNA extracted from bone marrow sample was used as a template. This assay utilized 999 hybrid capture oligonucleotides to sequence a 151.5kb panel of genes. The targeted region was circularised and amplified with Illumina adapters and sample specific barcodes. Sequencing was done on an Miseq sequencer (Illumina, USA) using the V2 500-cycle chemistry. The genes sequenced include ABL1-exon4-8, ASXL1-exon12-13, ASXL2-exon11-12, BCOR-exon4-15, BRAF-exon15, CALR-exon9, CBL- exon4,5,6,8 and 9, CSF3R-exon14-17, CUX1-exon15-23, DDX41-exons 1-17, DNMT3A-all exons, ETV6-exon1-8, EZH2-exon2-18, FLT3-exon14-16, 20, GATA1-exon2, GATA2-exon3-5, IDH1-exon4, IDH2-exon4, JAK2- exon11-21, KDM6A-exon2-29, KIT-exon2, 3, 7-18, KRAS-exon2-5, MPL-exon4-8 and 10-12, NF1-exon2-58, NPM1-exon 11, NRAS-exon2-5, PHF6-exon2-9, PTEN- exon 5-7, exon 9, PTPN11-exon 3, 13, 14, RAD21-exon 2-13, RUNX1-exon5-8, SAMD9 exon 3, SAMD9L exon 5, SETBP1-exon2-6, SF3B1-exon6-24, SH2B3-exon3-8, SMC1A-exon2-5,8-17,22,23, STAG2-exon5-30, TET2- exon3-11, TP53-allexons, U2AF1-exon2-7 and 11, U2AF2-exon5, 6, 11, WT1-exon 7, 11, ZRSR2-exon2-11. In addition, SRSF2 exons 1 and 2 were tested using tiled primers with amplicon based sequencing approach. We used the GATK best practices framework for identification of sequence variants. Sequencing data was analysed using a customized pipeline that incorporates adapter trimming using ea-utils, read self-assembly using PEAR, bwa (v0.7.12) for mapping to the human genome (build hg19/ GRCh37), samtools (v. 0.1.19) & GATK v3.8 for pre- processing the aligned file. Seven variant callers (MuTect, VarScan2, LoFreq, Strelka, VarDict, Freebayes and Platypus) were used to generate mutation calls. Finally, the variants were annotated with population frequency databases [1000G (African, Admixed American, East Asian, Finnish, Non-finnish European, South Asian), Exome Aggregation Consortium datasets, NHLBI-ESP (6500 genomes) as well as the COSMIC database (v80)] as well as the Cosmic database (Cosmic v83). Only exonic, non-synonymous, mis-sense, non-sense mutations and in-frame/frameshift indels were reported. The variants were reported in VAF% with their HGVS genomic / protein/transcript ID nomenclature, derived from the Variant Validator tool online in accordance with the ACMG & AMP guidelines and their clinical significance described using Varsome, ClinVar, NCBI-VEP and CosmicVar tools.
Wherever required, Immunophenotyping was performed using bulk lysis staining method and further acquired on 3-laser, 13-color Dx FLEX flowcytometer (Beckman Coulter, USA). Immunophenotyping data was analysed with Kaluza (v 2.1) software (Beckmann Coulter, USA), using a pre-defined template based approach.
Patients were stratified into various risk groups depending upon the clinical characteristics, cytopenias and/or blast count, abnormal karyotype as well as molecular profile as applicable, and treatment strategies evaluated and compared accordingly.
Results
Out of a total of sixty-one patients enrolled during the last two-year period, 6.56% had polycythemia vera (PV), 4.92% were essential thrombocythemia (ET), 3.3% had chronic eosinophilic leukemia, 3.3% has juvenile myelomonocytic leukemia (JMML) and 32.8% were diagnosed with primary myelofibrosis. Apart from this, 32.8% and 16.4% had MDS and chronic myelomonocytic leukemia (CMML) respectively (Figure 1).
Figure 1. Frequency % of Different MPNs, MDS, and MDS/MPN Overlap.
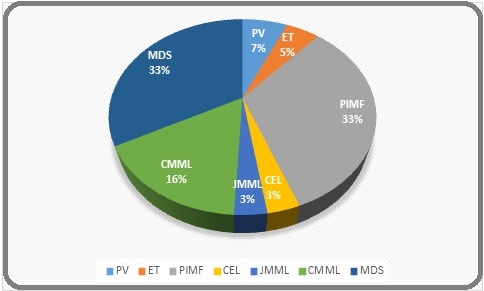
Other subtypes were not observed in our study.
In PIMF, 50% patients were JAK2- mutated while 30% were triple negative for JAK, CALR and MPL. The commoner epigenetic modifiers among MPNs were ASXL1, TET2 and IDH2. The predominant CMML molecular signatures in our patients were NRAS, U2AF2, SETBP1, ASXL and SH2B3 (detailed list attached in Supplementary Table). Interestingly, no karyotypic abnormalities were found in any of our MPN and MPN/MDS patients except presence of Monosomy 16 and Monosomy 7 in one patient of PIMF and JMML respectively. Figures 2 and 3 reveal the cytogenetic and molecular spectrum of these MDS patients.
Figure 2. Cytogenetic Abnormalities in MDS Patients.
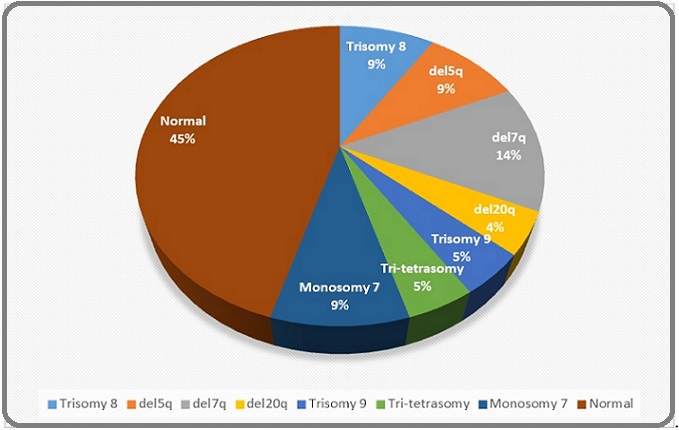
Figure 3. Frequency of Molecular Mutations in MDS Patients.
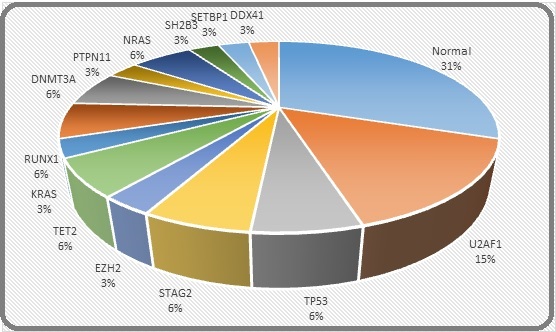
There was no significant effect of WHO changes and recently introduced molecular scoring models on the diagnosis and risk stratification of all these MPN and MDS/MPN patients except for the fact that JMML was shifted from MDS/MPN to MPN category. However, in MDS and PIMF patients, recent WHO subtyping plus IPSS-M & GIPSS scoring respectively enabled refining of risk groups, which will be elaborated below.
Among the twenty patients diagnosed with MDS, median age at presentation was 54 years with an age range of 15-71 years and M: F ratio of 1:1. Table 1 highlights the changes in the classification of same cohort of MDS patients (n=20) in accordance with 2017 and 2022 WHO revised guidelines respectively.
| 2017 WHO subtypes | 2022 WHO subtypes |
| MDS-single lineage dysplasia – 02 | MDS with defining genetic abnormalities- 05 |
| MDS-multi lineage dysplasia – 07 | MDS with low blasts and isolated 5q deletion-03 |
| MDS-excess blasts I -05 | MDS with low blasts and SF3B1 mutation-nil |
| MDS-excess blasts II -06 | MDS with biallelic TP53 inactivation-02 |
| MDS, morphologically defined- 15 | |
| MDS, hypoplastic 03 | |
| MDS with low blasts 07 | |
| MDS with increased blasts | |
| · MDS-IBI 02 | |
| · MDS-IBII 06 | |
| MDS with fibrosis 03 |
In our MDS patients, MIPSS-R and IPSS-R risk scores ranged from -1.5 to +2.29 and 1.0 to 8.5 respectively. As per the MIPSS-R scoring, 15% of the MDS patients were upstaged to high or very high categories, while 50% patients remained unmodified Table 2.
| Case No | CTG | NGS | R-IPSS CAT | IPSS-R Score | MIPSS-R | M SCORE -GROUP |
| 1 | Trisomy 8 | Normal | Very High | 6.5 | 1.37 | High |
| 2 | del (5q) and del (7q) | Normal | High | 6 | 0.3 | Moderate |
| 3 | del (20q); Monosomy 5/del (5q) | Normal | Low | 2 | -0.95 | Low |
| 4 | Normal | Positive for a missense mutation in exon 6 of U2AF1 gene, nonsense mutation in exon 9 of TP53 gene and a missense mutation in exon 8 of TP53 gene | Intermediate | 4.5 | 2.04 | Very High |
| 5 | del (20q) | Normal | Low | 2 | -1.2 | Low |
| 6 | del (5q) and del (7q) | Normal | High | 5 | 0.56 | High |
| 7 | Trisomy 8 | positive for substitution in STAG2 gene, substitution in exon 17 of EZH2 gene, multiple mutations in the TET2 gene: each in exon 3 and exon 7, substitution in exon 5 of TP53 gene and substitution in exon 3 of KRAS gene | Very High | 6.5 | 2.19 | Very High |
| 8 | Trisomy 9 and tri-tetrasomy 22 | substitution in exon 2 of U2AF1 gene and substitution in exon 25 of STAG2 gene | Very High | 6.5 | 1.56 | Very High |
| 9 | Monosomy 7 | non frameshift deletion in exon 5 of RUNX1 gene, substitution in exon 23 of DNMT3A gene, substitution in exon 3 of PTPN11 gene, substitution in exon 3 of NRAS gene and substitution in exon 3 of SH2B3 gene | High | 6 | 2.07 | Very High |
| 10 | Monosomy 7 | Normal | Very High | 8.5 | 1.51 | Very High |
| 11 | Normal | Normal | Very Low | 1 | -1.5 | Very Low |
| 12 | Normal | Positive for a missense mutation in the exon 6 of TET2 gene | Intermediate | 3.5 | -0.46 | Moderate |
| 13 | Normal | positive for substitution in exon 4 of SETBP1 gene, substitution in exon 2 of U2AF1 gene, substitution in exon 14 of DNMT3A gene | Intermediate | 6 | 1.65 | Very High |
| 14 | Normal | positive for substitution in exon2 of U2AF1 gene and substitution in exon 2 of NRAS gene. | High | 6 | 2.29 | Very High |
| 15 | Normal | Normal | Intermediate | 4 | 0.63 | High |
| 16 | Normal | positive for Substitution in exon 2 of U2AF1 gene and exon 7 of RUNX1 gene. | Intermediate | 3.5 | 1.28 | High |
| 17 | Normal | positive for substitution in exon 9 of DDX41 gene | High | 6 | 0.68 | High |
| 18 | Normal | Normal | Low | 2.5 | -0.64 | Low |
| 19 | del (7q) | Normal | Very High | 7 | 1.6 | High |
| 20 | Normal | Normal | Very High | 6.5 | 0.93 | High |
Among the twenty patients diagnosed with PIMF, median age at presentation was 55.5 years with an age range of 36-72 years and M: F ratio of 1:1. DIPSS score was applied in all patients and compared with GIPSS score based on presence of very high risk karyotype, unfavourable karyotype, absence of CALR Type I mutation, presence of ASXL1, SRSF2 or U2AF1Q157. DIPSS and GIPSS risk scores ranged from 0-4 and 0-2 respectively. As per the GIPSS scoring, 30% of the PIMF patients were down staged from high to intermediate or low categories, while the remaining 70% patients remained unaffected Table 3.
| Case No | CTG | NGS | GIPSS | DIPSS |
| 1 | Monosomy 16 | NORMAL | Int-1 | High |
| 2 | ND | Positive for two substitutions each in exon 2 and exon 6 of U2AF1 gene. | Int-2 | High |
| 3 | Normal | Normal | Int-1 | High |
| 4 | NA | Positive for missense mutation in exon 8 of CBL gene | Int-1 | High |
| 5 | Normal | Positive for substitution in exon 6 of SH2B3 gene, substitution in exon 6 of U2AF1 gene and frameshift deletion in exon 3 of ETV6 gene | Int-1 | High |
| 6 | Positive for a substitution in the exon 14 of JAK2 gene | Int-1 | High | |
| 7 | Normal | Positive for a missense mutation in exon 14 of JAK2 gene | Int-1 | Int-2 |
| 8 | Normal | Positive for substitution in exon 14 of JAK2 gene | Int-1 | Low |
| 9 | Normal | Positive for a substitution in exon 14 of JAK2 gene, a substitution in exon 12 of ASXL1 gene and a substitution in exon 10 of TET2 gene | Int-2 | Int-1 |
| 10 | Normal | Positive for substitution in exon 14 of JAK2 gene and a substitution in exon 4 of IDH2 gene | Int-1 | Int-1 |
| 11 | Normal | Positive for stopgain mutations in exon 35 of NF1 gene | Int-1 | Int-2 |
| 12 | Normal | Positive for a substitution in exon 14 of JAK2 gene and a substitution in exon 3 of TET2 gene | Int-1 | Int-1 |
| 13 | Normal | Positive for a 52bp deletion in exon 9 of the CALR gene & frameshift deletion in BCOR gene | Low | Int-2 |
| 14 | Normal | Positive for missense mutation in JAK2 gene | Int-1 | Int-1 |
| 15 | Normal | Positive for substitution in exon 14 of JAK2 gene | Int-1 | Low |
| 16 | Normal | Multiple substitutions in the exon 7 and exon 13 of EZH2 gene, frameshift insertion in the exon 9 of CALR gene (type 2) and substitution in exon 2 of NRAS gene | Low | Int-1 |
| 17 | Normal | Tested positive for complex delins in exon 10 of MPL gene, substitution in exon 4 of EZH2 gene, substitution in the exon 8 of CBL gene and substitution in the exon 2 of NRAS gene | Int-1 | Int-1 |
| 18 | Normal | Positive for substitution in exon 14 of JAK2 gene | Int-1 | Low |
| 19 | Positive for substitution in exon 14 of JAK2 gene | Int-1 | Low | |
| 20 | Normal | Positive for frameshift insertion in the exon 9 of the CALR gene (5bp insertion) | Low | Int-1 |
Discussion
Philadelphia negative MPNs, MDS/MPN and MDS unite a group of heterogeneous hematologic diseases that harbor a common set of specific genetic variations, often occurring as a result of stepwise accumulation of genetic alterations in the hematopoietic stem cells or progenitor cells. As per the 2022 revised WHO classification of myeloid neoplasms, few changes have been introduced in these entities, some of which have been discussed here. The term myelodysplastic neoplasms (abbreviated MDS) replaces myelodysplastic syndromes, stressing on their neoplastic nature. They have been grouped into those having defining genetic abnormalities and those that are morphologically defined. The distinction between single lineage and multi-lineage dysplasia is now considered optional and more emphasis has been placed on factors like fibrosis, hypoplasia and blast count. Among the MDS/MPNs entities, atypical CML has been renamed as MDS/MPN with neutrophilia and the previously incorporated entity, JMML has been shifted to the category of MPNs. MDS/MPN with ringed sideroblasts and thrombocytosis is re-defined based on SF3B1 mutation and renamed MDS/MPN with SF3B1 and thrombocytosis. The cut-off for monocytes in diagnosis of CMML has been reduced from 1x109/L to 0.5x109/L, in order to include the oligo-monocytic variants of CMML. CMML-0 subtype has been removed, CMML-MD and CMML-MP subtypes have been adopted and abnormal partitioning of peripheral blood monocytes (classical monocytes >94%) is introduced as a new supporting criteria. Philadelphia-negative MPNs have undergone minor changes in the form of omission of the qualifier NOS from CEL, inclusion of JMML and updating of its diagnostic criteria as well as removal of increased red cell mass as a diagnostic criterion from Polycythemia vera.
The development of next generation sequencing (NGS) techniques has helped to define the molecular landscape of myeloid neoplasms. Nowadays, myeloid NGS panels can be applied to perform a complete analysis of genetic alterations in a single approach. These alterations are mainly single-nucleotide variants and small insertions and deletions, although some myeloid gene panels also cover copy number variants (CNV) and translocations. Gene selection is an important issue that should be based on a thorough review of the literature. Clinical myeloid panels must include genes that are potentially “actionable”, i.e., genes whose variants are implicated in diagnosis, prognosis, treatment, and follow-up [5].
More than 90% of these patients harbor somatic mutations in a group of genes that is common across the spectrum of myeloid neoplasms Mutations recurrently affect epigenetic regulators (TET2, ASXL1, DNMT3A, EZH2, IDH2), splicing factors (SRSF2, SF3B1, U2AF1, ZRSR2), genes involved in signaling pathways (N/KRAS, CBL, JAK2, PTPN11, NF1, MPL,
SH2B3,ETNK1,CSF3R), transcription factors (RUNX1, GATA2, CUX1, ETV6) and cohesin complex components (STAG2.SMC1A, SMC3, RAD21) and others such as TP53, SETBP1 etc. Moreover, the development of specific pathway-targeting treatments in combination with advances in genomic biomarker discovery is paving the way for precision medicine for many patients [6-10]. The study by Palomo et al revealed that the most frequently occurring mutations in MDS patients are those that affect DNA methylation such as TET2, DNMT3A, followed by EZH2, ASXL1 and RNA splicing factors in decreasing order of frequency [7,8]. This is in striking contrast to our study where splicing factor mutation U2AF1 was the commonest occurring in MDS patients followed by DNMT3A and TET2. IDH1/2 mutations were not seen in our MDS patients.
Similarly, the study by Jian et al revealed that the most common molecular signatures in CMML were TET2 (60%) and ASXL1 (40%), followed by SRSF2 & RAS pathways [9]. But in our study, NRAS mutations were the commonest, followed by U2AF1 and SETBP2 mutations. Moreover, in our study, frequency of driver mutations remains unchanged but DNMT3A mutations were found in none whereas mutations involving EZH2, TET2, NRAS and U2AF1 predominated the molecular picture of our PIMF patients. This is again different from the molecular profiling of western patients [10].
The majority of MPN and MPN/MDS patients have a normal karyotype; however, around 25–30% present with clonal cytogenetic abnormalities. Common alterations include trisomy 8, abnormalities of chromosome 7, trisomy 21 and complex karyotypes [11]. These abnormalities are also present at different frequencies in other myeloid neoplasms such as MDS, MPN and AML. In our study, prevalence of abnormal karyotypic abnormalities across MDS/MPNs as well as MPNs was almost negligible as compared to western literature. This brings us to the impression that cytogenetics may not be compulsorily performed in these subset of patients, once bcr-abl fusion transcript has been ruled out by PCR techniques.
The standard of care in our subset of MDS patients was to treat very low to low risk patients with best supportive care in the form of transfusion support, antibiotics and growth factors, erythropoietin etc. High to very high risk patients were treated with more intensive regimens or hypomethylating agents till they achieve remission and then were counselled for allogenic bone marrow transplantation if eligible. Intermediate patients were triaged depending upon severity of cytopenias, blast percentage and TP53 mutation status to decide approach to therapy [12]. As per the IPSS-M applied in this study retrospectively, no change was required to be made in terms of therapeutic decision making in any of the patients. The reason behind this was patients in intermediate to high/ very high risk categories as per the IPSS-R scoring, were already on hypomethylating agents such as azacytidine or decitabine owing to concomitant TP53 mutation status or worsening cytopenias or high blast count etc.
In PIMF, observation alone was advised for low to very low risk patients. In intermediate risk patients or those not willing/eligible for transplant, conventional treatment was offered in the form of androgens, prednisone, thalidomide, and danazol for anemia and hydroxyurea and/or ruxolitinib for constitutional symptoms and symptomatic splenomegaly [4] while “very high” and “high” risk disease was counselled for allogenic stem cell transplantation. As per the GIPSS scoring applied retrospectively in PIMF patients, no intensification of therapy was required in any of the patients.
The above mentioned variations in cytogenetic and genomic profiling of our subset of Indian patients reaffirm the fact that the Indian population is very different at the genetic level from that of the West and may explain the reason why there was not much effect on treatment decisions in our subset of MDS and PIMF patients on the basis of MIPSS & GIPSS scores in comparison to R-IPSS and DIPSS Scores respectively.
In Conclusion, this brings us to the impression that molecular profiling helps in better risk stratification of patients across all groups as well as in making therapeutic decisions. However, in resource constrained settings as in the Indian subcontinent, it is not always possible to stratify patients on the basis of molecular signatures and hence, scoring models such as DIPSS and IPSS-R holds their ground strongly even today for offering appropriate therapy to patients without compromising on quality care. Moreover, due to financial constraints, definitive or targeted therapy such as ruxolitinib or allo-BMT may not be feasible in most patients and hence resorting to conservative measures is the only solution in this part of the world.
Acknowledgments
Statement of Transparency and Principals:
· Author declares no conflict of interest
· Study was approved by Research Ethic Committee of author affiliated Institute.
· Study’s data is available upon a reasonable request.
· All authors have contributed to implementation of this research.
References
- The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms Khoury JD , Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF , Bejar R, et al , Hochhaus Andreas. Leukemia.2022;36(7). CrossRef
- A novel scoring system integrating molecular abnormalities with IPSS-R can improve the risk stratification in patients with MDS GU S, Xia J, Tian Y, Zi J, Ge Z. BMC cancer.2021;21(1). CrossRef
- Molecular International Prognostic Scoring System for Myelodysplastic Syndromes Bernard E, Tuechler H, Greenberg PL , et al . NEJM Evid.2022;1(7). CrossRef
- Primary myelofibrosis: 2021 update on diagnosis, risk-stratification and management Tefferi A. American Journal of Hematology.2021;96(1). CrossRef
- Next-Generation Sequencing Improves Diagnosis, Prognosis and Clinical Management of Myeloid Neoplasms Carbonell D, Suárez-González J, Chicano M, Andrés-Zayas C, TriviñoJC , Rodríguez-Macías G, Bastos-Oreiro M, et al . Cancers.2019;11(9). CrossRef
- Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms Palomo L, Meggendorfer M, Hutter S, Twardziok S, Ademà V, Fuhrmann I, Fuster-Tormo F, Xicoy B, et al . Blood.2020;136(16). CrossRef
- Genetic Aspects of Myelodysplastic/Myeloproliferative Neoplasms Palomo L, Acha P, Solé F. Cancers.2021;13(9). CrossRef
- Genetics of MDS Ogawa S. Blood.2019;133(10). CrossRef
- Mutations in chronic myelomonocytic leukemia and their prognostic relevance Jian J., Qiao Y., Li Y., Guo Y., Ma H., Liu B.. Clinical & Translational Oncology: Official Publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico.2021;23(9). CrossRef
- Epigenetics in Myeloproliferative Neoplasms McPherson S, McMullin mf , Mills K. Journal of Cellular and Molecular Medicine.2017;21(9). CrossRef
- Cytogenetic evolution in myeloproliferative neoplasms with different molecular abnormalities Kim sy , Koo M, Park Y, Kim H, Choi Q, Song I, Jo D, Kim J, Kwon GC , Koo SH . Blood Cells, Molecules & Diseases.2019;77. CrossRef
- Treatment of MDS Platzbecker U. Blood.2019;133(10). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2023
Author Details