Epigenetics and Cancer: A Comprehensive Review
Download
Abstract
Cancer is a disease with extraordinary clinical significance, with much of medical research being devoted to it. Innumerable factors are relevant in fully understanding cancer but the epigenetic aspect stands out. Epigenetics is the study of changes, often germ-line, to the genome affecting the gene expression by silencing certain genes and modifying the gene expression. The three primary mechanisms for epigenetic changes are DNA methylation, histone modification and non-coding RNA (ncRNA) associated gene silencing. While epigenetics is a pivotal mechanism for the regular maintenance of a myriad of processes- including in cell differentiation and adaptability- aberrant epigenetic changes can lead to depreciated/altered gene function which may ultimately culminate in cancer. Consequently, the connection between epigenetics and cancer has been intensely studied over the past two decades and has generated substantial clinical data attesting to the efficacy of epigenetics as a viable approach to understand cancer progression or therapy. In this review, we look at the fundamental epigenetic principles, the changes in the epigenome which can often be a precursor to cancer, analyse the increasingly important role of epigenetics in decoding carcinogenesis, explore the latest advancements in use of epigenetics in cancer therapy and how the reversible nature of these epigenetic changes have changed the way we approach cancer therapy.
Introduction
Conrad Waddington coined the term Epigenetics in 1942 for describing heritable changes to the phenotype without corresponding changes in the genetic code [1, 2]. Over time, this definition has become more excursive and is today defined as ‘‘the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence’’ [3]. The complement of DNA methylation, histone modification and other epigenetic mechanisms are collectively referred to as the epigenome. Failure in the maintenance of thesemechanisms can result in improper activation or inhibition of certain important genes, leading to diseases, notably cancer.
Cancer is a cluster of diseases characterized by the dysregulation of important cellular pathways that are involved in the regulation of cell cycle, proliferation, differentiation, DNA repair and cell death. Cellular transformation, tumor initiation, tumor progression and metastasis are coordinated by a complex network of interactions involving aberrant genetic and epigenetic alterations resulting in cancer [4]. The etiology of cancer was long considered to be anomalies in the genome. But as it became evident that the genome is regulated via epigenetic mechanisms, it became clearer that epigenetics and cancer are intrinsically entwined. A dysregulated epigenetic alteration can affect various aspects of cancer biology such as aberrant transcription, cancer initiation, cancer progression, drug resistance and metastasis. But unlike genetic mutations, which are difficult to amend, altered epigenetic mechanisms are more feasible targets for therapy, owing to their dynamic and reversible nature. Epigenetics have great potential for cancer therapies and reprogramming that allows recovery of normal function of the affected genes [5]. These advantages have led to an exponential increase in epigenetic research in the field of oncology to combat cancer and hence making them very promising and therapeutically relevant targets.
Epigenetic mechanisms are essential for normal development and maintenance of tissue specific gene expression in mammals [6]. Precisely how the genotype of an individual will interact and respond to the environment is determined by epigenetics. Deregulation of epigenetic processes can lead to altered gene expression and malignant transformation.In 2022, “nonmutational epigenetic reprogramming’’ was includedas one of the hallmarks of cancer [7]. The initiation and progression of cancer, traditionally seen as a genetic disease, is now realized to involve epigenetic abnormalities along with genetic alterations. Given the importance of epigenetic regulation in cancers, the treatment targeting epigenetics is becoming an attractive strategy of cancer therapy. Epigenetic treatment may therefore benefit cancer patients as monotherapy and in combination with other standard cancer treatment regimen.Recent advancements in the rapidly evolving field of cancer epigenetics have shown extensive reprogramming of every component of the epigenetic machinery in cancer, including DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs, specifically microRNA expression [8]. The epigenetic deregulation leads to activation of oncogenes and/or inactivation of tumor suppressor genes (TSGs).
The complexity and multifactorial nature of cancer presents a great challenge for the diagnosis and treatment. The epigenetic basis of cancer development revolutionized the field of cancer genetics in the post- genomic era, providing new targets for cancer therapy [9]. In this review, we take a comprehensive look at the current understanding of the epigenetic mechanisms at work in normal mammalian cells and their comparative aberrations that occur during carcinogenesis. We also discuss the prospect of epigenetic cancer monotherapy or in combination epidrug therapy in various cancer treatments. We hope to shower light on the significance of epigenetics in development and treatment of cancer.
Epigenetic mechanisms in normal versus cancer cells
Epigenetic mechanisms are essential for normal mammalian growth, development and maintenance of gene expression.The epigenetic marks are tightly and interdependently connected and are essential for the normal development and the maintenance of cellular homeostasis and functions in adult organisms, particularly for X-chromosome inactivation in females, genomic imprinting, silencing of repetitive DNA elements, regulation of chromatin structure, and proper expression of genetic information [10]. Epigenetic mechanisms primarily regulate the compactness and accessibility of chromatin by altering the local dynamics of chromatin. The interplay of different epigenetic modifications regulates the way the mammalian genome manifests itself in different cell types, developmental stages and disease states, including cancer [11]. The distinct patterns of these modifications present in different cellular states serve as a guardian of cellular identity. These epigenetic modifications display unique properties and distribution patterns in different mammalian cells. The distinct combinatorial patterns of these modifications, collectively termed the epigenome, are key determinants of cell fate and gene activity [12].
The well-balanced and precise epigenomic landscape of normal cells undergoes extensive distortion in cancer cells. Cancer cells emerge from normal healthy cells during the complex, multistep and progressive processof malignant transformation that involves both genetic and epigenetic aberrations [13]. It is characterized by the accumulation of multiple cancer-specific heritable phenotypes, including persistent proliferative signaling, resistance to cell death, evasion of growth suppression, replicative immortality, inflammatory response, deregulation of energy metabolism, genomic instability, induction of angiogenesis, and activation of invasion ultimately resulting in metastasis [14,15]. The complexity and the frequency of the epigenetic changes seen in cancer cells relies on multiple factors that results from myriad genetic mutations and environmental inputs which perturb the manifold nodes of epigenetic regulations (Figure 1).
Figure 1. Epigenetic Regulation in Cancer. Epigenetic marks change during foetal development (thus, turning mechanisms on/off), through adult life (through various exposures), and occur throughout the aging process. Epigenetics are influenced by a number of external and internal exposures, such as physical activity, diet, ageing and environment. These exposures lead to epigenetic alterations in our chromosome and thus causing genomic instability and promoting cancer.
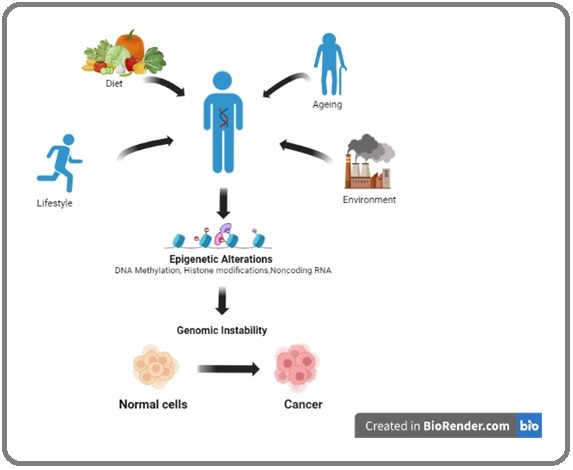
The cancer epigenome is characterized by global alterations in DNA methylation, histone modifications and non-coding RNA (ncRNA) mediated regulation of gene expression, which confers growth advantage to tumor cells and hence promotes cancer development [16, 2].
The Epigenome and its Elements
The epigenome is an umbrella term for a myriad of chemical compounds that influence gene expression by latching on to DNA and silencing or activating genes without altering the DNA sequence (Figure 2).
Figure 2. Overview of Epigenetic Mechanisms of Gene Expression. Nucleosomes are formed by DNA wrapped around a histone octamer that allows DNA to be condensed into chromatin and finally chromosomes. The regulation of chromatin structure occur through various post translational histone modifications such as, acetylation, phosphorylation, ubiquitiylation, methylation. All these histone modification results inweakening of histone interaction with DNA to increase chromatin accessibility. DNA methyltransferases (DNMT) methylate CpG islands near transcription start (promoter) sites inhibiting gene expression by impeding transcription factor binding to DNA. The regulation of transcription also occurs via non-coding RNA.
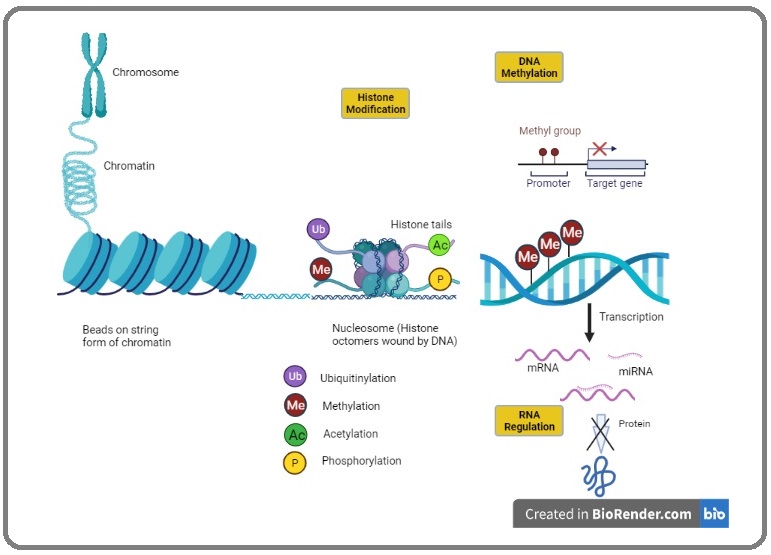
The epigenome is of paramount importance to understanding diseases such as cancer because it has the significant ability to change how genes are expressed in a cell. Every cell of an individual has essentially the same genome in its nucleus. But different cells of the body are specialized for different functions in different tissues or organs. The different functions are determined by when and how the different sets of genes are turned on or off [17]. The epigenome, containing a variety of chemical compounds, can bind to the DNA and may direct the genes to turn on or off. The identity of a cell is established and maintained by the physical accessibility of DNA in a chromatin. The organization of accessible DNA of a chromatin over the genome reflects a network of acceptable physical interactions between promoters, enhancers, insulators and chromatin-binding factors to regulate gene expression [18]. The epigenome mediated change in gene expression is influenced by a multitude of factors including environmental ones. For instance UV radiation has a known propensity of increasing the risk formelanoma and basal cell carcinoma and that is owing to the fact that the radiation influences the epigenome into causing changes to gene expression which ultimately results in cancer [19].
DNA Methylation
DNA methylation is a well-established epigenetic marker, in which a methyl group attaches itself in the C5 position in cytosine rings in CpG islands to form 5-methylcytosine (m5c) [20]. This additional methyl group directly influences gene expression by recruiting proteins involved in the gene expression of that particular gene and by inhibiting the affinity of transcription factors towards binding sites in DNA [21]. Promoter hypermethylation of the CpG islands of tumor suppressor genes is a major event in the initiation of many cancers and can affect genes involved in regulating the cell cycle, DNA repair, cell-to-cell interactions, apoptosis and angiogenesis pathways that are involved in cancer development [22-24]. The addition of methyl groups in cytosine is catalyzed by enzymes called DNA methyltransferases (DNMTs). DNMTs have three active types: DNMT1 maintains existing methylation patterns and DNMT3a and DNMT 3b are de novo methyltransferases [25]. During embryonic cell division, DNMT3a and DNMT3b are responsible for the differentiation of cells into specific cell types, by silencing the DNA and preventing certain proteins from being expressed. Following differentiation, DNMT1 is responsible to maintain this methylation during cell division and thereafter as well. DNA methylation occurs in the cytosine and modifies the particular CpG. In most adult cells, CpG sites are methylated to maintain chromosomal stability [26], however, at promoter regions, there are stretches of DNA having higher concentration of CpG sites, known as CpG islands that are generally not methylated in normal cells[27]. The promoter regions in the DNA contain regulatory elements that control the transcription of genes. In cancer though, the highly regulated methylation and demethylation is unbalanced, which leads to the hypermethylation of these promoter CpG islands, associated with the inactivation of TumorSuppressor Genes (TSGs).Global genomic DNA hypomethylationsare generally found in several tumor types and eventually lead to genomic instability [28-31].
According to the widely acknowledged Knudson’s two hit hypothesis, tumorigenesis is caused by mutations in and/or silencing of the two alleles of a tumor suppressor gene [32]. In case of a genetic history of cancer, one of the alleles is already mutated and the second can be silenced by DNA methylation while if no history of cancer exists both the alleles need to be methylated for carcinogenesis to occur. This is also why genetic history of cancer makes it far more likely for it to occur down the line. This further exemplifies the consequential role DNA methylation plays in initiating cancer [33]. DNA methylation also often leads to and is indicative of the Warburg Effect, a hallmark of cancer in which cells consume significantly higher amounts of glucose and perform glycolysis at higher rates and preferentially produce lactose, even in the presence of adequate oxygen [34].
In a large, systemic study, DNA methylations’ pivotal role has become clearer, with a 2018 study elucidating the role of the mechanism in 24 different types of cancers. It was found to be an important factor in a variety of cancers and often being responsible for various subtypes within a tumor. For instance, hypermethylation was found to be correlated to EBV (Epstein-Barr Virus) infection in stomach cancers, IDH1 and IDH2 mutations in glioma etc. Methylation was also found to be even more crucial in pediatric cancers since they typically consist of a constellation of epigenetic modifications rather than genetic drivers [35].
In recent times, hydroxymethylation has also been implicated in the development of cancer. It is well known that 5-methylcytosine (5mC) is oxidized to 5-hydroxymethylcytosine (5hmC) and other derivatives by methylcytosine dioxygenases TETs (Ten-Eleven Translocation family) in an active demethylation process [36-39]. Oxidation of 5mC leads to active DNA demethylation, while a certain proportion of 5hmC remains in the genome and serves as a regulatory function [40-44].Although 5hmCs contribute to the flexibility of chromatin and protect bivalent promoters from hypermethylation, the direct effect of 5hmCs on gene transcription is not known. Reduced expression of TET gene and lower hydroxymethylation levels have been reported inseveral human cancers [45-49].
Histone Modification
Another important epigenetic mechanism of gene expression are histone modifications. Histones are protein octamers that primarily provide structural support to DNA, making it possible for long DNA strands to fit inside cell nuclei by giving them a compact shape. The DNA strands are tightly coiled around the histones and therefore are incapable of undergoing gene transcription. The tight coiling is maintained due to DNA being a negatively charged polymer and histone being positively charged. In order for gene expression to occur the DNA needs to unwind and that can occur only when histones are modified (acetylated/methylated). This modification entails lysine residues of the N-Terminal extruding from the histone core being modified, thereby regulating access to the DNA for transcription [50-52]. These acetylation reactions are catalyzed by enzymes referred to as histone acetyltransferases (HATs) (which acetylates the lysine amino acid by transferring a molecule of acetyl group from acetyl-COA to convert the lysine into ε-N-acetyl lysine).
Aberrant epigenetic histone modifications have been implicated in pathogenesis of cancer and thereby impact clinical outcomes [53].More than 20 HATs have been identified, which are classified into 5 families-GNAT1, MYST, TAFII250, P300/CBP, and nuclear receptor coactivators such as ACTR.1. Histone deacetylases (HDACs) are responsible for catalyzing the hydrolytic removal of acetyl groups present in histone lysine residues. Currently, four classes of HDACs have been identified, class I, II, III and IV [54]. An imbalance in this process has been associated with tumorigenesis and cancer progression. Histones can be modified by phosphorylation, methylation, acetylation, ubiquitination etc. Post-transcriptional modificational errors in histones may lead to alteration in gene expression, which in turn may lead to cancer [55-56].
Histone methylation occurs mainly on the side of chains and arginines. Unlike acetylation, it does not alter the charge of the histone protein [57]. It is defined as the transfer of one, two, or three methyl groups from S-adenosyl-L-methionine to lysine or arginine residues of histone proteins by histone methyltransferases (HMTs). They control and regulate DNA methylation through transcriptional repression or activation.
Histone acetylation is the addition of an acetyl group such as -COCH. It is extensively involved in the regulation of cellular processes including chromatin dynamics and transcription, gene silencing, cell cycle progression, differentiation, and DNA replication [58]. Unlike DNA methylation, histone modification can cause activation or repression of genes, depending upon the residues modified and the type of modifications present. Specific patterns of histone modifications are present within distinct cell types that play a major role in determining cellular identity. For example, embryonic stem cells possess active (H3K4me3) and repressive (H3K27me3) marks as promoters of developmentally important genes [59].
Histone phosphorylation is controlled by two types of enzymes, having opposing modes of action. Kinases remove phosphate groups and phosphatases remove the phosphates. Three functions of these histones are known, which are, DNA damage repair, control of chromatin compaction during mitosis and meiosis, and regulation of transcriptional activity [60]. Histone phosphorylation establishes a platform for interactions with other histone modifications as it works in conjunction with them.
Non-coding RNA Modification
Non-coding RNAs are a cluster of RNAs that do not encode functional proteins but they play an important role in epigenetic control of gene expression.Among the epigenetic elements, non-coding RNAs (ncRNA) have received special attention recently, owing to theirrole in several epigenetic mechanisms controlling gene expression. This control is exerted at the level of chromatin structure modulation; transcriptional regulation and posttranscriptional modification [61]. ncRNAs operate as a key regulator of physiological programs in developmental and disease contexts,especially in cancer. They have been identified as oncogenic drivers and tumor suppressors in every major cancer type. Non-coding RNAs take up 70% of human genomes and based on the size, have been categorized into micro RNAs (miRNAs) with approximately 20 nucleotides and long non-coding RNAs (lncRNAs) with >200 nucleotides [62,63]. 50% of miRNAs arelocated in chromosomal regions that are prone to structural changes [64]. Numerous miRNAs have been shown to be dysregulated in various cancers including colorectal cancer and leukemia [65, 66]. lncRNAs have also been reported to have aberrant expression in various cancer and can act as either oncogenic or tumor suppressor factor, making them a potential diagnostic biomarker and therapeutic targets.
Regulation of gene expression in cancer progression
Gene expression is the process of transcription of a particular gene from DNAinto mRNAand then translation of mRNA to the amino acids and proteins which manifest as the phenotype. Alteration in gene expression is the primary factor for progression of the vast majority of human diseases, including cancer.Cancer is a multistep process associated with the accumulation of numerous molecular alterations, which impact cellular machinery within the tumor and its microenvironment. Numerous genetic alterations (mutations, loss of heterozygosity, etc.) have been associated with carcinogenesis and have been shown to behave as cancer driver mutations [67]. These alterations result in aberrant gene expressions.
Normal cells turn neoplastic by tumorigenesis, a complicated, multi-stage molecular process. These tumors may then later turn malignant, giving rise to cancer. Of the particular processes which influence carcinogenesis in gene expression, microRNAs are particularly noteworthy. They are a type of non-coding RNA of roughly 22 nucleotides. These miRNAs, along with theargonaute protein form the base of the RNA interference (RNAi) mechanism, which is a major form of gene silencing [68]. The RNAi molecules were discovered by Dr. Fire and Dr.Mello [69] while working on C. elegans, who later went on to win the Nobel Prize in Physiology and Medicine for this discovery. RNAi is a standard method of transcriptional regulation in eukaryotes, protecting not only against exogenous genes but also against endogenous genes with potential to grow aberrantly such as transposons. These molecules have also been known to be involved in cell growth control, which is often a gene which plays a central part in carcinogenesis. As a result, this mechanism has a propensity/potential for targeting cancer-related genes and therefore has been a newfound target for cancer therapies [70-72].
Two models have been presented to explain how a tumor progresses: (i) the cancer stem cell (CSC) model, which regards stem cells as the origin of oncogenic transformation; (ii) the clonal evolution model, which suggests that the initial oncogenic change is acquired progressively by non CSCs, and there is no doubt that epigenetic changes are involved in both the models.
DNA methylation acts as a regulator of gene expression by switching genes “on” and “off”. Hypermethylation of promoters in CpG islands is the most recognized mechanism of epigenetic alterations in cancer cells and has been found to be an important factor in various cancer types. Alterations of both genome-wide and gene-specific DNA methylation are critical events in tumor initiation and progression (Figure 3).
Figure 3. Aberrant DNA Methylation Contributes to Tumorigenesis (A). Hypermethylation of the promoter of tumor suppressor genes (TSGs) or gene family leads to its silencing and contributes to tumorigenesis.(B) Hypomethylation of oncogene promoters leads to their activation and contributes to tumorigenesis. (C) Genome-wide DNA hypomethylation may lead to chromosomal instability or cell differentiation, which contributes to tumorigenesis [76].
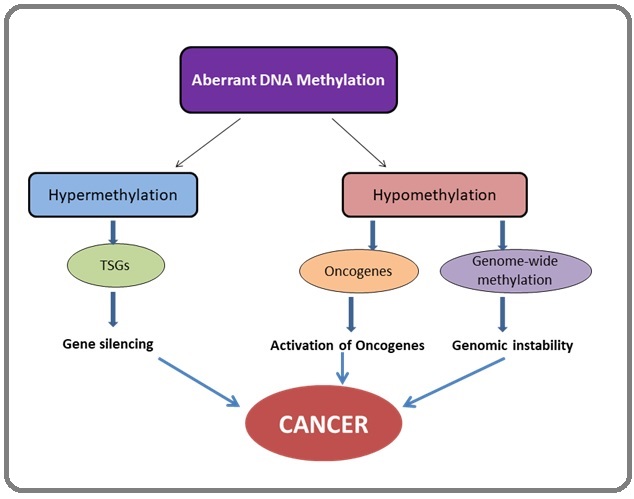
Abundant tumor suppressor genes are under hypermethylation like RASSF10 in renal cancer and SIX3 in glioblastoma [73, 74]. Apart from DNA methylation, unbalanced histone modification is abused by cancers that adhere to the CSC model. Essentially, all the aberrations caused by the modification of histones have been known to occur at individual gene promoters, due to inappropriate targeting of these enzymes.
Epigenetic alterations in some common cancers
Gastric cancer
Epigenetic hallmarks, along with genetic aberrations, have been identified in different subgroups of gastric cancer (GC). Accumulating evidence suggests that epigenetic abnormalities in GC are not mere bystander events, but rather promote carcinogenesis through active mechanisms [76]. To date, aberrant DNA methylation is the most extensively studied deregulated epigenetic mechanism in GC [77]. For instance, known tumor suppressors or tumor-related genes (p16, RUNX3, MLH1, CDH1, etc.) are silenced by promoter methylation in GC and its precancerous lesions [78].
Colorectal cancer
The pathophysiology of colorectal cancer (CRC) is driven by varying genetics and epigenetic alterations. A substantial number of patients develop chemoresistance and hence face treatment failure. This leads to a high mortality rate for advanced disease. The study of epigenetic changes has bought to light the missing link between certain CRC-specific gene expression patterns and the absence of genetic alterations over past two decades. These insights, among others, have not only improved our understanding of CRC pathophysiology but have also opened the door to the discovery of new disease biomarkers and therapeutic targets. Aberrant promoter hypermethylation silences tumor suppressor genes and global DNA hypomethylation causes genomic instability, leading to CRC initiation and progression [79]. The potential prognostic utility of DNA methylation in CRC has been increasingly highlighted. For instance, CpG island methylator phenotype (CIMP), characterized by a high prevalence of promoter CpG island methylation, have been demonstrated to be an independent prognostic factor in CRC [80]. Hypermethylation of IGFBP3 was associated with poor CRC prognosis in few separate studies [81, 82].
Breast cancer
The epigenetics of breast cancer is relatively well characterized and plays an important role in cancer development. Epigenetic drivers are found early in breast cancer carcinogenesis, including early DNA methylation and alteration in chromatin states [83]. Furthermore, lncRNAs and miRNAs have been found to affect a large numbers of genes- regulating their function and driving carcinogenesis and the development of intratumoral heterogeneity. Throughout the evolution of breast cancer, epigenetic reprogramming of these tumors occurs and has been found on the APC, CDH1 and CTNNB1 genes [84]. Distinct patterns of reprogramming have been found through from ductal hyperplasia, ductal carcinoma in-situ to invasive carcinoma. Interestingly, the finding of distinct methylation patterns is not just confined to malignant breast cancer cells [85]. Adjacent, histopathologically normal tissue has been found to demonstrate similar DNA methylation profiles to cancerous cells, revealing a field effect. Epigenetic profiling of tumors and circulating DNA has raised the possibility that measurement of DNA methylation may be used prognostically. So far, however, few of these have made it to clinical practice and/or trials of epigenetic alterations to guide cancer therapy [86, 87]. As with other tumor types, HDAC inhibitors have been trialed in breast cancer, with varying degrees of success and at present, there is a lack of convincing placebo-controlled data from a large trial to support routine clinical use [84]. Although breast tumors are also frequently hypomethylated on a genome-wide scale, the number of genes reported to be hypomethylated in breast cancer is relatively small [88].
When assessing epigenetic changes, both DNA and histone level modifications must be examined, particularly if the modified genes are therapeutic targets.
Prostate cancer
Epigenetic dysregulation, including changes in DNA methylation, histone modifications and nucleosome remodeling, occurs at every phase of prostate cancer (PCa) development and progression. These aberrations are responsible for silencing tumor-suppressor genes, activating oncogenic drivers, and driving therapy resistance. Targeting epigenetic pathways is an emerging therapeutic strategy in prostate cancer. EZH2, the catalytic unit of polycomb repressive complex (PRC2) responsible for H3K27me3 and gene repression, has been identified as a promising target in PCa [89]. In addition, overexpression of DNMT in PCa has also been documented. Till now, more than 50 genes with common aberrant hypermethylation have been identified in prostate cancer [90]. These genes encompass many cellular functions including cell cycle control, apoptosis, hormone response, DNA repair and damage prevention, signal transduction, tumor invasion and tumor suppression. Frequent promoter methylation of genes such as APC, CCND2, GSTP1, RARβ2, RASSF1A and PTGS2 has been observed in PCa [91].
Lung cancer
Lung cancer involves genetic and epigenetic events in the respiratory epithelium. While somatic genetic aberrations, such as mutations and copy number alterations, play a well-known role in carcinogenesis, epigenetic alterations are more frequent than somatic mutations in lung cancer. Recent advances in epigenetics provide a better understanding of the underlying mechanism of carcinogenesis. DNA hypermethylation is a hallmark in lung cancer and an early event in carcinogenesis. Many of the epigenetic events in lung cancer affect cancer hallmarks, such as proliferation, invasion, metastasis, apoptosis, and cell cycle regulation [92]. In addition to cancer hallmarks, several important signaling pathways are affected by epigenetic deregulation in lung cancer, such as the ERK family, the NF-kB signaling pathway, and the Hedgehog signaling pathway [93]. Epigenetic events also provide insight into the discovery of putative cancer biomarkers for early detection, disease monitoring, prognosis, risk assessment, and oncotherapy.
Epigenetic pathways linked to carcinogenesis and cancer progression
Epigenetic changes are present in all human cancers and are known to cooperate with genetic alterations to drive the cancer phenotype. During carcinogenesis genes can become activated in a way that either enhances cell growth or prevents cell death, known as the oncogene. Also, genes can become inactive and are unable to stop these processes of uncontrolled cell growth; these are the tumor-suppressor genes. Interplay between these results in the formation of cancer.
There are three ways by which tumor suppressor genes can become inactive: (1) through mutations disabling their functions; (2) a gene maybe lost and is not available to work; (3) a gene can be switched off in a somatic heritable fashion by epigenetic changes, rather than by mutation of the DNA sequence.
It is extensively established that the standard epigenetic mechanisms are subject to global disruption during tumorigenesis. Changes in these epigenetic mechanisms also notably involve the hypermethylation of CpG islands and hypomethylation of the broader genome. The hypermethylation of these CpG islands is particularly associated with silencing of certain tumor suppressor genes and genes otherwise related to cell growth, so much so that various studies have demonstrated that promoter-related CpG islands are hypermethylated in tumor cells. Instances include the BRCA1 gene in breast carcinoma, in which healthy tissue’s CpG islands was found to be unmethylated in every case, but methylation being present in 13% of the cases [94]; in case of Glutathione S-transferase P (GSTP1) in prostate cancers [95], healthy CpG islands were found to be entirely unmethylated but cancerous tissue showing methylation in roughly 90% of the cases, establishing a strong connection between CpG methylation and carcinogenesis [96].
Epigenetic modifications can also drastically alter the treatment plan or prognosis for certain cancers, such as in Diffuse Midline Gliomas, if the H2K27 histone mutation is detected, it worsens the prognosis significantly and renders it incurable and inoperable [97].
The Polycomb Repressive Complex 2 (PRC2), particularly EZH2, an enzymatic component of PRC2 methylates the H3K27 and H3K9 site. EZH2 is widely considered to be a key epigenetic regulator of cancer progression and has been, as a result, been extensively studied. It has been found to have been aberrantly overexpressed in primary tumors in prostate cancer, breast cancer, and ovarian cancer. EZH2 has also been found to increase tumor growth and if ectopically expressed it has been found to be a growth factor for fibroblasts and leads to cellular proliferation resulting in malignant cancer [98, 99].
A newer pathway of regulated cell death (RCD), ferroptosis, has also been shown to have epigenetic causes [100]. Ferroptosis, is a novel form of regulated cell death mediated by iron-dependent lipid peroxidation, has been receiving increasing attention since its discovery in 2012. Owing to the highly iron-dependent physiological properties of cancer cells, targeting ferroptosis is a promising approach in cancer therapy.
Long non-coding RNA (lncRNA) are a group of RNA made up of more than 200 nucleotides, yet have minimal protein coding capacity. Due to their capacity of interacting directly with DNA, RNA and proteins they influence a myriad of crucial cell processes including cell differentiation and, notably, RCD and ferroptosis. In non- small cell lung cancers (NSCLC) lncRNAs suppress the development of NSCLCs by ferroptosis’ pathways [101]. Epigenetics also plays an intriguing role in the breast cancer progression via CAFs (Cancer Affected Fibroblasts). CAFs exhibit a shift towards pyruvate and lactate production. (With this glycolytic shift itself being sustained by epigenetic reprogramming of HIF-1α). The hypoxia induced by this prompt the epigenetic revision of normal fibroblasts, converting them into CAF-like transcriptomes, which promotes breast cancer progression [102].
Epigenetic therapy in cancer
As the role of epigenetics in cancer became clearer, treatments targeting epigenetic changes in cancer came to the forefront. Drugs specifically targeting these epigenetic changes, referred to as epidrugs, target cancer by changing the epigenetic modifications in chromatin and DNA by regulating the enzymes required for the upkeep of transcriptional modifications and reactivating various epigenetically silenced genes including the ones involved in DNA repair and tumor suppression. They do so primarily by inhibiting Histone Deacetylase and DNA Methyltransferases, thereby reversing and reducing their potential impact in further cancer progression.
Epigenetic factors not only fuel tumorigenesis and cancer progression, but are often implicated in certain cancers evolving drug resistance by epigenetic changes and therefore serve as a prime target for further improving prognosis.
Cancer cells often exhibit alterations in DNA methylation. With focal DNA hypermethylation at CpG-rich sites and genome-wide hypomethylation. It is associated with the silencing of tumor suppressor genes and activation of oncogenes. Targeting DNMTs to restore the epigenome of cancerous cells to a ‘normal’ state has been proven to be an effective way for cancer therapy. Researchers have classified two categories of DNMT inhibitors – Nucleoside analogs and non-nucleoside. With a modified cytosine ring, nucleoside analogs can be included in newly synthesized DNA and covalently bound to DNMTs. This inhibits them from transferring methyl groups to hemimethylated DNA of progeny cells, leading to generalized demethylation and proteasomal degradation of DNMTs.
One instance is targeting ERα (Estrogen Receptor Positive) positive breast cancers via endocrine therapy. The therapy has been in use for several decades but in recent years it was found that in a few early stage cancers and almost all metastasized ones, resistance to the endocrine therapy had evolved. This was owing to loss-of-function epigenetic changes in ARID1A, the gene controlling endocrine therapy sensitivity. Currently, no FDA approved epidrug exists to target this [103].
A number of epidrugs have been approved by the United States Food and Drug Administration (FDA), including Azacitidine and Decitabine, targeting DNMTi for the treatment of Acute Myeloid Leukemia; Belinostat, targeting HDACi in peripheral T cell Lymphomas; Panobinostat, Bortezomib and Dexamethasone targeting HDACi in Multiple myeloma (Table 1).
| S.N. | Group | Compound | Target | Treated Cancer | Reference |
| 1 | DNMT Inhibitor | 5-azacytidine | DNMT1 | AML, MDS | 107 |
| 5-aza - 2/- Deoxycytidine | DNMT1 | AML, MDS | 108 | ||
| Decitabine | DNMT1 | MDS, Breast Cancer | 109, 110 | ||
| 5GL-110 (Gaudecetabine) | DNMT1 | AML, MDS | 111 | ||
| RX-3117 | DNMT1 | Pancreatic Cancer | 112 | ||
| MG-98 | DNMT1 | DMS, AML, Renal Cancer | 113 | ||
| 2 | HDAC Inhibitor | Vorinostat | HDAC1,2,3 & 6 | CTCL | 114 |
| Romidepsin | HDAC1 & 2 | CTCL, PTCL | 115 | ||
| Pracinostat | Class I,II & IV HDAC | AML | 116 | ||
| Abexinostat | HDAC1,2,3,6,8 & 10 | Relapsed Lymphoma | 117 | ||
| Panobinostat | pan - HDAC | Multiple Myeloma | 118 | ||
| Entinostat | HDAC1,9 & 11 | Hodgkin Lymphoma, Breast Cancer | 119 | ||
| 3 | HAT Inhibitor | Anacardic Acid | P 300, PCAF& Tip60 | Breast Cancer | 120 |
| C646 | p300 | AML, Prostate Cancer | 121 | ||
| 4 | HMT Inhibitor | EPZ-5676 | DOTIL | Haematological Malignancies | 122 |
| Tazemetostat | EZH2 | Lymphoma, Solid Tumors | 123 | ||
| AZ505 | SMYD2 | Glioma | 124 | ||
| 5 | HDM Inhibitor | Tranylcypromine Analogue | LSD1 | AML | 125 |
| Pargyline | LSD1 | Prostate Cancer | 126 | ||
| 6 | BET Inhibitor | OTX015 | BRD2,3&4 | Leukemia | 127 |
| CPI-0610 | Dem amino-Terminal Bromodomains (BD1 and B) | Relapsed/ Refractory Lymphoma | 128 | ||
| 7 | PARP Inhibitor | Olaparib | PARP1 | Ovarian, Breast Cancer | 129, 130 |
| Veliparib | PARP1 | Breast, NSCLC | 129, 130 |
Out of the seven FDA approved epidrugs, only two target DNMTs among them the latest being Azacitidine and Decitabine. 5-Azacitidine (azacytidine) and 5-aza- 2′-deoxycytidine (decitabine) are the two most studied nucleoside analogs. They induce methylation and reactivate silenced genes in human cancers.Azacytidine functions as ribonucleoside and preferentially incorporates into RNA through phosphorylation. However, it can be incorporated into DNA to a lesser extent through the ribonucleotide reductase pathway. Unlike azacytidine, decitabine is a deoxyribose analog that incorporates directly into DNA following phosphorylation, thus inducing DNA demethylation at much lower concentrations than azacytidine.Both of these drugs have received FDA approval for the treatment of Acute Myeloid Leukemia (AML), chronic myelomonocytic leukemia (CMML), and myelodysplastic syndrome (MDS) [104].
Multiple phase three studies of these two drugs have depicted improved response rates and an overall increase in survival when compared to standard therapies, especially in older AML patients. Although these drugs are very effective in targeting cancer cells, they suffer from poor chemical and metabolic stability and high toxicity. Given the success and drawbacks of azacytidine and decitabine, the prodrugs of decitabine or azacytidine have been developed as next-generation DNMT inhibitors, which exhibit superior pharmacokinetic profiles.Sinceepidrugs are specific to the presence of particular epigenetic errors, they are also a gateway to an era of increasingly personalized medicine, with a particular error entailing treatment with a tailor made epidrug [9].
Epigenetic drugs in combination with other therapies
The integration of epigenetic therapies with standard cancer treatment, including chemotherapy, immunotherapy and targeted therapy has emerged as an attractive cancer therapy regimen (Figure 4) and supports the hypothesis that epi-drugs can synergize with other anticancer agents and reverse cancer therapy resistance in preclinical models [105].
Figure 4. The Integration of Epigenetic Medicines with other Standard Chemotherapy, Targeted Therapies, and Immunotherapy .
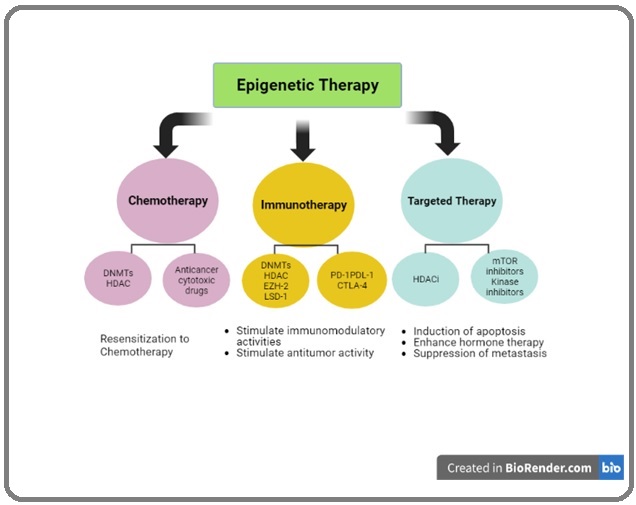
An underlying theme is illustrated by data linking chemoresistance to epigenetic events in cancer stem-like cells. The studies implicated an HDM; JARID1, which decreases the active transcription mark H3K4me3; and low doses of HDAC have reversed the stem cell-like properties and chemoresistance. A similar HDM, JARID2, may also drive stem-like human melanoma cells [106]. Low doses of DNMTis can likewise inhibit cancer stem cell properties, enhance apoptosis, and block entry into the cell cycle. These examples illustrate the potential for broad reprogramming of cancer cells with epigenetic therapies to prime them for subsequent therapies.
Epigenetic drugs in combination with Chemotherapy
An appealing strategy for preventing chemoresistance is coupling epigenetic agents with chemotherapeutic drugs that cause DNA damage has emerged [107-109]. Chemoresistance is typically coupled with epigenetic programming, such as aberrant methylation of DNA and alterations in histone acetylation, which can be restored by DNMT and HDAC inhibitors [110-112]. This aberrant DNA is associated with methylation of key genes in mTOR signalling/AKT/PTEN/PI3K pathway which promotes resistance in various solid tumors via alteration in cell survival, motility, apoptosis, angiogenesis, cell metabolism and cell proliferation [113].
Epigenetic drugs in combination with Immunotherapy
Immune checkpoint therapies, involved in boosting anticancer immune responses by preventing checkpoint molecule interaction, have resulted in a significant advancement in cancer treatment [114-116]. Even though immunoglobulins against checkpoint proteins such as CTLA-4, programmed deathligand 1 (PD-L1) and programmed cell death protein 1 (PD-1 or CD279) exhibited promising anticancer effects, their therapeutic use may be restricted due to poor antigen presentation and inadequate T-cell response. These limitations can be overcome by immunomodulatory actions driven by epigenetic remodelling [117-119]. Inhibiting epigenetic regulators such as lysine-specific demethylase 1A (LSD1), enhancer of zeste homolog 2 (EZH2), HDAC and DNMT elicit several immunomodulatory actions in cancer cells, including overexpression of MHC class I molecules, tumor antigens, and PD-1 ligands [120]. Knocking out such epigenetic proteins also initialises the production of endogenous retroviral components and double-stranded RNA in cancer cells, that also activates interferon signalling, helps to stimulate antineoplastic T-cell immunity, and makes cancer cells more susceptible to checkpoint blockade therapies [121].
Epigenetic drugs in combination with Targeted therapy
The development of targeted treatments, which utilize chemicals intended to specifically interact with certain mutant/aberrantly signaling proteins, has constituted a real shift in cancer therapy paradigm [122]. Pharmacological treatment of mutant kinases produces fast clinical outcomes in genetically specified populations [123, 124]. Resistance against targeted therapy, on the other hand, is almost unavoidable. Genetic changes and transcriptional reprogramming are two resistance mechanisms that can be reversed using epigenetic treatments [125]. Epigenetic alterations are involved in oncogenesis of NSCL but its role in EGFR-TKI resistance is still unclear. However, HDAC inhibition was reported to overcome tolerance to a variety of kinase inhibitors. For example, a relatively novel oral histone deacetylase inhibitor MPT0E028 was able to rise apoptosis induced by the first-line epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) erlotinib in EGFR-TKI resistant NSCLC cells [126]. A preliminary study reported that combining EGFR- TKIs and vorinostat reversed EGFR-TKI resistance and promoted apoptosis in NSCLC cell models [127]. HDAC inhibitors have also been shown to reverse the mammalian target of rapamycin (mTOR) TKI resistance in a number of malignancies as it has been observed that methylation of genes impact mTOR signalling pathway due to epigenetic alteration [128, 129].
Challenges and Future prospects
Epigenetic modifications are among the first aberrations to occur in cancer development. Targeting these epigenetic changes has been demonstrably effective in tackling cancer. But, despite the use of epidrugs in cancer therapy having several clinical advantages, there are some challenges that need to be considered and worked upon.
Even after combination therapy, the reversible nature of methylation persists, which may lead to re-methylation and re-silencing of tumor suppressor genes. The toxicity and instability of a few epidrugs (e.g. 5 aza C and 5 aza D) may complicate their use in clinical settings under varying physiological conditions. Furthermore, treatment may result in the activation of typically silenced genes due to lack of specificity of methylating agents and this may contribute to tumorigenesis.
As is clear from extensive research, epigenetic therapies have achieved remarkable clinical responses in hematologic malignancies. Even so, their activity as monotherapies has been less encouraging in some solid tumors due to limited efficacy and/or unfavorable toxicity. The reason may potentially be that these solid tumors arise from more terminally differentiated cells and are inherently less sensitive to epigenetic perturbation, and therefore prolonged inhibition of the epigenetic targets and optimizing dosing schedules may improve the efficacy. In addition, most clinical trials of early generation epigenetic drugs including DNMT and HDAC inhibitors have followed a one-size-fits-all approach; the therapeutic efficiency and utility of which has been largely restricted due to the lack of appropriate patient selection with transcriptional or epigenetic signatures. Thus, unless our understanding of the epigenome continues to improve and the key regulators of epigenetic control are identified, therapies targeting epigenetic modifications within the human genome will remain of limited value and not see fruition in the form of widespread dissemination in the clinical setting.
Nevertheless, several unanswered questions remain. One area of future research within the field of cancer epigenetics is epigenetic heterogeneity in cancer. Heterogeneity of cancer cells within a given cancer mass is a well-recognized feature of cancer and remains to be one of the reasons that development of effective anti-cancer therapeutics is challenging. Although certain epigenetic therapies have demonstrated promising clinical outcomes and have received regulatory approvals in hematologic malignancies, it remains challenging to achieve and maintain the therapeutic efficacy in solid tumors. Appropriate patient selection and optimizing trial design and dosing schedules may improve their clinical efficacy. Importantly, epigenetic drugs in conjunction with other therapies such as targeted or immune-based therapies may offer the best opportunity to augment clinical responses in solid tumors.
In conclusion, cancer being multifactorial in nature poses tremendous challenge in its diagnosis and treatment. The given utility of dynamic and reversible epigenetic regulation in cancer has revolutionized the field of cancer development and hence making it an attractive target for cancer therapy.
As the role of epigenetics has come to the forefront in cancer research in the last few decades, it has given rise to an entirely new branch of understanding of cancer as a whole, upending and adding on to long held perceptions in tumorigenesis and treatment. We have discussed the myriad of epigenetic mechanisms, including DNA methylation and Histone Modifications and how they pertain and contribute to the deregulation of the epigenetic landscape thereby becoming a factor leading to carcinogenesis. We have also looked at a range of epigenetic point mutations which can often serve as the primary drivers of certain cancers.
Notably, we have elucidated how the specific epigenetic mechanism can be targeted to serve as a base for developing an entirely new class of cancer treatments: Epidrugs. We have also analysed at depth, epidrugs that have been approved and those that show promising preliminary results in clinical trials. In addition, we have discussed the utility and potential of epigenetic biomarkers having gone into detail for the ones that hold the most promise.
We firmly believe that as much attention that the connection between epigenetics and cancer has garnered in the past decades, it still has much to offer in our understanding of cancer particularly, in the development of new therapies and biomarkers which will optimistically prove to be detrimental in reducing the stubbornly flat cancer death rate across the globe.
Even though more precise mechanisms must be researched, it is commonly known that epigenetic activities are critical in both normal biochemical mechanisms and tumor pathways, and that epigenetic status is often greatly changed during cancer onset. As a result, epigenometargeted therapy appears to be a potential option for cancer therapy. Because of the complexities of cancer, epigenetic changes have affected a range of cancer properties, including oncogene expression along with tumor repressor genes, as well as cell signalling, that results in rapid cancer growth, infiltration, and metastases. There are several strategies and advancement to target epigenetic machinery. These include combination therapy,adjuvant therapy, and probiotics, CRISPR Cas-9 editing, phytochemicals, Phototherapy, cold atmospheric plasma, nanocarriers and terahertz rays. All these novel therapeutic strategies lead to tackle drug resistance along with better treatment option with high rate of cure and recovery. Epigenetic targeting appears to be a potential anticancer therapeutic method based on the results achieved. Many features of cancer onset are associated with epigenomes. It is required to have a deeper knowledge of the exact processes behind such modifications in various cancers. Meanwhile, improved therapy approaches, including a range of combinations, have yet to be developed. Epigenetic modifications lead to chemoresistance. Hence it is necessary to consider epigenetic machinery while treating cancer. Terahertz rays and cold atmospheric plasma are novel targets and it is the need of hour to perform more research on these novel tools. Bacteria based therapy or probiotics is also very hot topic since 2020 in the field of oncology that is why their adjuvant and combination therapeutics would gain much attention in the future with high cure rate. More work is required on phytochemicals and CRISPR based therapy to deal with epigenetic alterations in cancer.
Acknowledgements
N/A
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AP was a major contributor in writing the manuscript. ST was a major contributor in writing the manuscript.
SP provided critical feedback on the manuscript that was pivotal in its suitability for peer review.
Abbreviations
DNA- Deoxyribonucleic Acid; RNA- Ribonucleic Acid; CpG - 5’—C—phosphate—G—3’; DNMT - DNA Methyltransferases; EBV - Epstein-Barr virus; IDH1/2 - Icocitrate Dehydrogenase ½; HAT - Histone Acetyltransferases; Acetyl-CoA - Acetyl Coenyzme A; GNATs - Gcn5-related N-acetyltransferases; MYST - MOZ-Ybf2 (Sas3)-Sas2-Tip60; TAF II 250 - TBP- associated factors ; TBP - TATA-binding proteins; CBP
- CREB-binding protein; CREB - cAMP-response element binding protein; HDAC - HitoneDeacetyltranferases; HMT
- Histone Methyltransferases; PRMT5 - Protein arginine methyltransferases; mRNA- Messenger Ribonucleic Acid; miRNA- Micro Ribonucleic Acid; RNAi- Ribonucleic acid interference; C. Elegans- Caenorhabditis elegans; CSC- Cancer Stem Cell model; BRCA1- Breast cancer gene 1; GSTP1- Glutathione S-Transferase P ; PRC2- Polycomb Repressive Complex 2; RCD- Regulated Cell Death; lncRNA- Long Non-Coding RNA; NSCLC- Non- small cell lung cancers; CAFs- Cancer affected fibroblasts; ERα- Estrogen receptor positive; FDA- Food and Drug Administration; AML- Acute Myeloid Leukemia; CTCL - Cutaneous T-cell lymphoma; CMML- Chronic Myelomonocytic Leukemia; MDS- Myelodysplastic syndrome; VIM - Vimentin; CRC - Colorectal Cancer; BPH - Benign Prostate Hypersplasia; RARbeta2 - Retinoic acid receptor beta 2; RASSF1A - Ras association domain family protein1 isoform A
References
- A decade of exploring the cancer epigenome - biological and translational implications Baylin SB , Jones PA . Nature Reviews. Cancer.2011;11(10). CrossRef
- Harnessing the potential of epigenetic therapy to target solid tumors Ahuja N, Easwaran H, Baylin SB . The Journal of Clinical Investigation.2014;124(1). CrossRef
- Genes, genetics, and epigenetics: a correspondence Wu Ct N, Morris JR . Science (New York, N.Y.).2001;293(5532). CrossRef
- Genomic and Epigenomic Alterations in Cancer Chakravarthi BVSK , Nepal S, Varambally S. The American Journal of Pathology.2016;186(7). CrossRef
- Epigenetic Determinants of Cancer Baylin SB , Jones PA . Cold Spring Harbor Perspectives in Biology.2016;8(9). CrossRef
- Epigenetics in cancer Sharma S, Kelly TK , Jones PA . Carcinogenesis.2010;31(1). CrossRef
- Hallmarks of Cancer: New Dimensions Hanahan D. Cancer Discovery.2022;12(1). CrossRef
- Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Signal Transduction and Targeted Therapy.2019;4. CrossRef
- Epidrugs: targeting epigenetic marks in cancer treatment Miranda Furtado CL , Dos Santos Luciano MC , Silva Santos RD , Furtado GP , Moraes MO , Pessoa C. Epigenetics.2019;14(12). CrossRef
- Epigenetic alterations in cancer Ilango S, Paital B, Jayachandran P, Padma PR , Nirmaladevi R. Frontiers in Bioscience (Landmark Edition).2020;25(6). CrossRef
- Epigenetic modifications: basic mechanisms and role in cardiovascular disease Handy DE , Castro R, Loscalzo J. Circulation.2011;123(19). CrossRef
- Epigenetic modification regulates tumor progression and metastasis through EMT (Review) Tan T, Shi P, Abbas MN , Wang Y, Xu J, Chen Y, Cui H. International Journal of Oncology.2022;60(6). CrossRef
- Hallmarks of cancer: the next generation Hanahan D, Weinberg RA . Cell.2011;144(5). CrossRef
- Revisiting the hallmarks of cancer Fouad YA , Aanei C. American Journal of Cancer Research.2017;7(5).
- The hallmarks of cancer: a long non-coding RNA point of view Gutschner T, Diederichs S. RNA biology.2012;9(6). CrossRef
- Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy Lu Y, Chan Y, Tan H, Li S, Wang N, Feng Y. Molecular Cancer.2020;19(1). CrossRef
- Chromatin accessibility and the regulatory epigenome Klemm SL , Shipony Z, Greenleaf WJ . Nature Reviews. Genetics.2019;20(4). CrossRef
- The DNA-encoded nucleosome organization of a eukaryotic genome Kaplan N, Moore IK , Fondufe-Mittendorf Y, Gossett AJ , Tillo D, Field Y, LeProust EM , Hughes TR , Lieb JD , Widom J, Segal E. Nature.2009;458(7236). CrossRef
- Patterns of Ultraviolet Radiation Exposure and Skin Cancer Risk: the E3N-SunExp Study Savoye I, Olsen CM , Whiteman DC , Bijon A, Wald L, Dartois L, Clavel-Chapelon F, Boutron-Ruault M, Kvaskoff M. Journal of Epidemiology.2018;28(1). CrossRef
- DNA methylation in early development Geiman TM , Muegge K. Molecular Reproduction and Development.2010;77(2). CrossRef
- Effects of cytosine methylation on transcription factor binding sites Medvedeva YA , Khamis AM , Kulakovskiy IV , Ba-Alawi W, Bhuyan MSI , Kawaji H, Lassmann T, Harbers M, Forrest ARR , Bajic VB . BMC Genomics.2014;15(1). CrossRef
- Gene silencing in cancer in association with promoter hypermethylation Herman JG , Baylin SB . The New England Journal of Medicine.2003;349(21). CrossRef
- Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome Weber M, Hellmann I, Stadler MB , Ramos L, Pääbo S, Rebhan M, Schübeler D. Nature Genetics.2007;39(4). CrossRef
- Cancer epigenomics: DNA methylomes and histone-modification maps Esteller M. Nature Reviews. Genetics.2007;8(4). CrossRef
- DNA methyltransferases, DNA damage repair, and cancer Jin B, Robertson KD . Advances in Experimental Medicine and Biology.2013;754. CrossRef
- CpG islands and the regulation of transcription Deaton AM , Bird A. Genes & Development.2011;25(10). CrossRef
- CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function Jang HS , Shin WJ , Lee JE , Do JT . Genes.2017;8(6). CrossRef
- DNA methylation, methyltransferases, and cancer Robertson KD . Oncogene.2001;20(24). CrossRef
- Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1 Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, Kurihara H. Developmental Biology.2008;313(1). CrossRef
- Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Brothman AR , Swanson G, Maxwell TM , Cui J, Murphy KJ , Herrick J, Speights VO , Isaac J, Rohr LR . Cancer Genetics and Cytogenetics.2005;156(1). CrossRef
- Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer Hon GC , Hawkins RD , Caballero OL , Lo C, Lister R, Pelizzola M, Valsesia A, et al . Genome Research.2012;22(2). CrossRef
- Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview Wang L, Wu C, Rajasekaran N, Shin yk . Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology.2018;51(6). CrossRef
- The second hit of DNA methylation Di Ruscio A, Welner rs , Tenen dg , Amabile G. Molecular & Cellular Oncology.2016;3(3). CrossRef
- How DNA methylation affects the Warburg effect Zhu X, Xuan Z, Chen J, Li Z, Zheng S, Song P. International Journal of Biological Sciences.2020;16(12). CrossRef
- Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Cell Reports.2018;25(4). CrossRef
- Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1 Tahiliani M, Koh kp , Shen Y, Pastor WA , Bandukwala H, Brudno Y, Agarwal S, Iyer lm , Liu dr , Aravind L., Rao A. Science (New York, N.Y.).2009;324(5929). CrossRef
- The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain Kriaucionis S, Heintz N. Science (New York, N.Y.).2009;324(5929). CrossRef
- Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA He YF , Li BZ , Li Z, Liu P, Wang Y, Tang Qi, Ding J, et al . Science (New York, N.Y.).2011;333(6047). CrossRef
- Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine Ito S, Shen L, Dai Q, Wu SC , Collins LB , Swenberg JA , He C, Zhang Y. Science (New York, N.Y.).2011;333(6047). CrossRef
- The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes Gu TP , Guo F, Yang H, Wu H, Xu G, Liu W, et ak . Nature.2011;477(7366). CrossRef
- Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine Hackett JA , Sengupta R, Zylicz JJ , Murakami K, Lee C, Down TA , Surani MA . Science (New York, N.Y.).2013;339(6118). CrossRef
- Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells Pastor WA , Pape UJ , Huang Y, Henderson HR , Lister R, Ko M, McLoughlin EM , et ak . Nature.2011;473(7347). CrossRef
- Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation Ficz G, Branco MR , Seisenberger S, Santos F, Krueger F, Hore TA , Marques CJ , Andrews S, Reik W. Nature.2011;473(7347). CrossRef
- Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome Yu M, Hon GC , Szulwach KE , Song C, Zhang L, Kim A, Li X, et al . Cell.2012;149(6). CrossRef
- Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers Haffner MC , Chaux A, Meeker AK , Esopi DM , Gerber J, Pellakuru LG , Toubaji A, et al . Oncotarget.2011;2(8). CrossRef
- TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/β-catenin signaling pathway to suppress EMT in pancreatic tumor cells Wu J, Li H, Shi M, Zhu Y, Ma Y, Zhong Y, et al . Journal of experimental & clinical cancer research: CR.2019;38(1). CrossRef
- TET genes: new players in DNA demethylation and important determinants for stemness Mohr F, Döhner K, Buske C, Rawat VPS . Experimental Hematology.2011;39(3). CrossRef
- The genomic landscape of nasopharyngeal carcinoma Lin D, Meng X, Hazawa M, Nagata Y, Varela AM , Xu L, Sato Y, et al . Nature Genetics.2014;46(8). CrossRef
- Genome-wide characterization of 5-hydoxymethylcytosine in melanoma reveals major differences with nevus Salgado C, Oosting J, Janssen B, Kumar R, Gruis N, Doorn R. Genes, Chromosomes & Cancer.2020;59(6). CrossRef
- Changing chromatin fiber conformation by nucleosome repositioning Müller O, Kepper N, Schöpflin R, Ettig R, Rippe K, Wedemann G. Biophysical journal.2014;107(9). CrossRef
- Fast signals and slow marks: the dynamics of histone modifications Barth TK , Imhof A. Trends in Biochemical Sciences.2010;35(11). CrossRef
- Characterization of human epigenomes Wang Z, Schones DE , Zhao K. Current Opinion in Genetics & Development.2009;19(2). CrossRef
- Role of novel histone modifications in cancer Shanmugam MK , Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, Wang LZ , et al . Oncotarget.2018;9(13). CrossRef
- A short guide to histone deacetylases including recent progress on class II enzymes Park SY , Kim JS . Experimental & Molecular Medicine.2020;52(2). CrossRef
- Epigenetic histone code and autoimmunity Dieker J, Muller S. Clinical Reviews in Allergy & Immunology.2010;39(1). CrossRef
- Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome Elsheikh SE , Green AR , Rakha EA ., Powe DG , Ahmed RA , Collins HM , Soria D, et al . Cancer Research.2009;69(9). CrossRef
- Histone modifications and their role in epigenetics of atopy and allergic diseases Alaskhar Alhamwe B, Khalaila R, Wolf J, Bülow V, Harb H, Alhamdan F, Hii CS , et al . Allergy, Asthma, and Clinical Immunology: Official Journal of the Canadian Society of Allergy and Clinical Immunology.2018;14. CrossRef
- Histone Modifications and Cancer Audia je , Campbell rm . Cold Spring Harbor Perspectives in Biology.2016;8(4). CrossRef
- Histone Modifications in Stem Cell Development and Their Clinical Implications Völker-Albert M, Bronkhorst A, Holdenrieder S, Imhof A. Stem Cell Reports.2020;15(6). CrossRef
- Histone phosphorylation: a chromatin modification involved in diverse nuclear events Rossetto D, Avvakumov N, Côté J. Epigenetics.2012;7(10). CrossRef
- Circulating long non-coding RNAs in cancer: current status and future perspectives Qi P, Zhou xy , Du X. Molecular Cancer.2016;15(1). CrossRef
- Landscape of transcription in human cells Djebali S, Davis ca , Merkel A, Dobin a, Lassmann T, Mortazavi A, Tanzer A, et al . Nature.2012;489(7414). CrossRef
- Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets Chandra Gupta S, Nandan Tripathi Y. International Journal of Cancer.2017;140(9). CrossRef
- Origins and Mechanisms of miRNAs and siRNAs Carthew RW , Sontheimer EJ . Cell.2009;136(4). CrossRef
- Downregulated microRNAs in the colorectal cancer: diagnostic and therapeutic perspectives Hernández Rosa, Sánchez-Jiménez E, Melguizo C, Prados J, Rama AR . BMB reports.2018;51(11). CrossRef
- Epigenetics and Epi-miRNAs: Potential markers/therapeutics in leukemia Memari F, Joneidi Z, Taheri B, Aval SF , Roointan A, Zarghami N. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie.2018;106. CrossRef
- Targeting Loss of Heterozygosity: A Novel Paradigm for Cancer Therapy Zhang X, Sjöblom T. Pharmaceuticals (Basel, Switzerland).2021;14(1). CrossRef
- Molecular mechanisms of RNA interference Wilson RC , Doudna JA . Annual Review of Biophysics.2013;42. CrossRef
- Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C.. Nature.1998;391(6669). CrossRef
- MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions Catalanotto C, Cogoni C, Zardo G. International Journal of Molecular Sciences.2016;17(10). CrossRef
- MicroRNA-145-5p modulates Krüppel-like factor 5 and inhibits cell proliferation, migration, and invasion in nasopharyngeal carcinoma Yuan CH , Hsu WC , Huang AM , Yuan BC , Chen IH , Hsu CA , Chen RF , et al . BMC molecular and cell biology.2022;23(1). CrossRef
- RNA interference and its role in cancer therapy Mansoori B, Sandoghchian Shotorbani S, Baradaran B. Advanced Pharmaceutical Bulletin.2014;4(4). CrossRef
- RASSF10 is frequently epigenetically inactivated in kidney cancer and its knockout promotes neoplasia in cancer prone mice Richter AM , Woods ML , Küster MM , Walesch SK , Braun T, Boettger T, Dammann RH . Oncogene.2020;39(15). CrossRef
- The EGFR-ZNF263 signaling axis silences SIX3 in glioblastoma epigenetically Yu Z, Feng Ji, Wang W, Deng Z, Zhang Y, Xiao L, Wang Z, et al . Oncogene.2020;39(15). CrossRef
- Mutual regulation of microRNAs and DNA methylation in human cancers Wang S, Wu W, Claret FX . Epigenetics.2017;12(3). CrossRef
- How to stomach an epigenetic insult: the gastric cancer epigenome Padmanabhan N, Ushijima T, Tan P. Nature Reviews. Gastroenterology & Hepatology.2017;14(8). CrossRef
- DNA methylation as a molecular biomarker in gastric cancer Tahara T, Arisawa T. Epigenomics.2015;7(3). CrossRef
- DNA and histone methylation in gastric carcinogenesis Calcagno DQ , Gigek CO , Chen ES , Burbano RR , Smith MDAC . World Journal of Gastroenterology.2013;19(8). CrossRef
- Aberrant DNA methylation profiles of inherited and sporadic colorectal cancer Sahnane N, Magnoli F, Bernasconi B, Tibiletti MG , Romualdi C, Pedroni M, Ponz de Leon M, et al . Clinical Epigenetics.2015;7. CrossRef
- Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis Juo Y. Y., Johnston F. M., Zhang D. Y., Juo H. H., Wang H., Pappou E. P., Yu T., Easwaran H., Baylin S., Engeland M., Ahuja N.. Annals of Oncology: Official Journal of the European Society for Medical Oncology.2014;25(12). CrossRef
- IGFBP-3 Gene Methylation in Primary Tumor Predicts Recurrence of Stage II Colorectal Cancers Fu T, Pappou EP , Guzzetta AA , Calmon MDF , Sun L, Herrera A, Li F, et al . Annals of Surgery.2016;263(2). CrossRef
- IGFBP3 methylation is a novel diagnostic and predictive biomarker in colorectal cancer Perez-Carbonell L, Balaguer F, Toiyama Y, Egoavil C, Rojas E, Guarinos C, Andreu M, et al . PloS One.2014;9(8). CrossRef
- Epigenetics and environment in breast cancer: New paradigms for anti-cancer therapies Thakur C, Qiu Y, Fu Y, Bi Z, Zhang W, Ji H, Chen F. Frontiers in Oncology.2022;12. CrossRef
- Cancer epigenetics in solid organ tumours: A primer for surgical oncologists Drake tm , Søreide K. European Journal of Surgical Oncology: The Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology.2019;45(5). CrossRef
- Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, et al . Genes & Diseases.2018;5(2). CrossRef
- DNA Methylation Cancer Biomarkers: Translation to the Clinic Locke WJ , Guanzon D, Ma C, Liew YJ , Duesing KR , Fung KYC , Ross JP . Frontiers in Genetics.2019;10. CrossRef
- Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction Nassar FJ , Msheik ZS , Nasr RR , Temraz SN . Clinical Epigenetics.2021;13(1). CrossRef
- The epigenetics of breast cancer Jovanovic J, Rønneberg JA , Tost J, Kristensen V. Molecular Oncology.2010;4(3). CrossRef
- Epigenetics in prostate cancer treatment Jones K, Zhang Y, Kong Y, Farah E, Wang R, Li C, Wang X, et al . Journal of Translational Genetics and Genomics.2021;5. CrossRef
- Epigenetics in breast and prostate cancer Wu Y, Sarkissyan M, Vadgama JV . Methods in Molecular Biology (Clifton, N.J.).2015;1238. CrossRef
- Advances in Prognostic Methylation Biomarkers for Prostate Cancer Lam D, Clark S, Stirzaker C, Pidsley R. Cancers.2020;12(10). CrossRef
- Current Landscape of Epigenetics in Lung Cancer: Focus on the Mechanism and Application Shi YX , Sheng DQ , Cheng L, Song XY . Journal of Oncology.2019;2019. CrossRef
- Signal transduction in cancer Sever R, Brugge JS . Cold Spring Harbor Perspectives in Medicine.2015;5(4). CrossRef
- Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors Esteller M., Silva J. M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., et al . Journal of the National Cancer Institute.2000;92(7). CrossRef
- Detailed methylation analysis of the glutathione S-transferase pi (GSTP1) gene in prostate cancer Millar D. S., Ow K. K., Paul C. L., Russell P. J., Molloy P. L., Clark S. J.. Oncogene.1999;18(6). CrossRef
- The DNA methylation landscape in cancer Skvortsova K, Stirzaker C, Taberlay P. Essays in Biochemistry.2019;63(6). CrossRef
- Diffuse Midline Gliomas With Histone H3 K27M Mutation in Adults and Children: A Retrospective Series of 164 Cases Zheng L, Gong J, Yu T, Zou Y, Zhang M, Nie L, Chen X, et al . The American Journal of Surgical Pathology.2022;46(6). CrossRef
- Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Biomarker Research.2018;6. CrossRef
- EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer Bracken AP , Pasini D, Capra M, Prosperini E, Colli E, Helin K. The EMBO journal.2003;22(20). CrossRef
- Ferroptosis: The Silver Lining of Cancer Therapy Tang Z, Huang Z, Huang Y, Chen Y, Huang M, Liu H, Ye QA , Zhao J, Jia B. Frontiers in Cell and Developmental Biology.2021;9. CrossRef
- The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu S, Tao Y. Molecular Cancer.2020;19(1). CrossRef
- Epigenetic Reprogramming of Cancer-Associated Fibroblasts Deregulates Glucose Metabolism and Facilitates Progression of Breast Cancer Becker LM , O'Connell JT , Vo AP , Cain MP , Tampe D, Bizarro L, Sugimoto H, et al . Cell Reports.2020;31(9). CrossRef
- Epigenetic mechanisms in breast cancer therapy and resistance Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL , Morey L. Nature Communications.2021;12(1). CrossRef
- Clinical advances in targeting epigenetics for cancer therapy Feng S, De Carvalho DD . The FEBS journal.2022;289(5). CrossRef
- Oral Azacitidine (CC-486) for the Treatment of Myelodysplastic Syndromes and Acute Myeloid Leukemia Cogle CR , Scott BL , Boyd T, Garcia-Manero G. The Oncologist.2015;20(12). CrossRef
- Decitabine/Cedazuridine: First Approval Dhillon S. Drugs.2020;80(13). CrossRef
- Epigenetics in myelodysplastic syndromes Heuser M, Yun H, Thol F. Seminars in Cancer Biology.2018;51. CrossRef
- Epigenetic Therapies and Biomarkers in Breast Cancer Brown LJ , Achinger-Kawecka J, Portman N, Clark S, Stirzaker C, Lim E. Cancers.2022;14(3). CrossRef
- Guadecitabine (SGI-110): an investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia Daher-Reyes GS , Merchan BM , Yee KWL . Expert Opinion on Investigational Drugs.2019;28(10). CrossRef
- RX-3117 (fluorocyclopentenyl cytosine): a novel specific antimetabolite for selective cancer treatment Balboni B, El Hassouni B, Honeywell RJ , Sarkisjan D, Giovannetti E, Poore J, Heaton C, et al . Expert Opinion on Investigational Drugs.2019;28(4). CrossRef
- MG98, a second-generation DNMT1 inhibitor, in the treatment of advanced renal cell carcinoma Amato RJ , Stephenson J, Hotte S, Nemunaitis J, Bélanger K, Reid G, Martell RE . Cancer Investigation.2012;30(5). CrossRef
- Vorinostat in cutaneous T-cell lymphoma Duvic M, Vu J. Drugs of Today (Barcelona, Spain: 1998).2007;43(9). CrossRef
- Role of human nucleoside transporters in the uptake and cytotoxicity of azacitidine and decitabine Damaraju VL , Mowles D, Yao S, Ng A, Young JD , Cass CE , Tong Z. Nucleosides, Nucleotides & Nucleic Acids.2012;31(3). CrossRef
- A real-world study of panobinostat, weekly bortezomib and dexamethasone in a very heavily pretreated population of multiple-myeloma patients Bird S, Pawlyn C, Nallamilli S, Sriskandarajah P, Kaiser M, Yong K, Popat R, Rabin N, Boyd K. British Journal of Haematology.2020;191(5). CrossRef
- Pracinostat plus azacitidine in older patients with newly diagnosed acute myeloid leukemia: results of a phase 2 study Garcia-Manero G, Abaza Y, Takahashi K, Medeiros BC , Arellano M, Khaled SK , Patnaik M, et al . Blood Advances.2019;3(4). CrossRef
- Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight Ho TCS Terence C. S., Chan AHY , Ganesan A.. Journal of Medicinal Chemistry.2020;63(21). CrossRef
- Recent advances in epigenetic anticancer therapeutics and future perspectives Ren L, Yang Y, Li W, Yang H, Zhang Y, Ge B, Zhang S, Du G, Wang J. Frontiers in Genetics.2022;13. CrossRef
- Emerging roles of anacardic acid and its derivatives: a pharmacological overview Hemshekhar M, Sebastin Santhosh M, Kemparaju K, Girish KS . Basic & Clinical Pharmacology & Toxicology.2012;110(2). CrossRef
- Histone Demethylase GASC1 Inhibitor Targeted GASC1 Gene to Inhibit the Malignant Transformation of Esophageal Cancer through the NOTCH-MAPK Signaling Pathway Li Q, Qu B, Shen H, Deng H, Sun L. Annals of Clinical and Laboratory Science.2022;52(2).
- The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis Gao Y, Ge W. Cell Death & Disease.2018;9(2). CrossRef
- EZH2: a novel target for cancer treatment Duan R, Du W, Guo W. Journal of Hematology & Oncology.2020;13(1). CrossRef
- The histone methyltransferase SMYD2 is a novel therapeutic target for the induction of apoptosis in ovarian clear cell carcinoma cells Kojima M, Sone K, Oda K, Hamamoto R, Kaneko S, Oki S, Kukita A, et al . Oncology Letters.2020;20(5). CrossRef
- Installation of Pargyline, a LSD1 Inhibitor, in the HDAC Inhibitory Template Culminated in the Identification of a Tractable Antiprostate Cancer Agent Ojha R, Chen IC , Hsieh CM , Nepali K, Lai RW , Hsu KC , LinTE , et al . Journal of Medicinal Chemistry.2021;64(24). CrossRef
- LSD1/KDM1A inhibitors in clinical trials: advances and prospects Fang Y, Liao G, Yu B. Journal of Hematology & Oncology.2019;12(1). CrossRef
- BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells Coudé MM , Braun T, Berrou J, Dupont M, Bertrand S, Masse A, Raffoux E, et al . Oncotarget.2015;6(19). CrossRef
- A Phase I Study of Pelabresib (CPI-0610), a Small-Molecule Inhibitor of BET Proteins, in Patients with Relapsed or Refractory Lymphoma Blum KA , Supko JG , Maris MB , Flinn IW , Goy A, Younes A, et al . Cancer Research Communications.2022;2(8). CrossRef
- PARP inhibitors in cancer therapy: promise, progress, and puzzles Ellisen LW . Cancer Cell.2011;19(2). CrossRef
- Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action Pommier Y, O'Connor MJ , Bono J. Science Translational Medicine.2016;8(362). CrossRef
- Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Nature Reviews. Clinical Oncology.2020;17(2). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2023
Author Details