Association of NQO1 C609T (Pro187Ser) with Risk of Oral Submucous Fibrosis in Eastern Indian Population
Download
Abstract
Objective: Oral submucous fibrosis (OSF) is a debilitating disease mainly attributed to chewing areca nut with a 7.4-13% malignant transformation rate. The present study explores the role of NADPH quinone oxidoreductase 1 (NQO1) C609T (Pro187Ser) polymorphism in susceptibility to OSF among habitual areca nut chewers in an eastern Indian population.
Material and Methods: In this hospital-based study, 152 controls and 179 OSF cases were genotyped at NQO1 C609T polymorphic site by PCR-RFLP and its effect on NQO1 expression in OSF tissue was studied to determine the risk of the disease.
Results: Overall, about 18% of the total OSF cases were detected carrying minor TT alleles (Ser/Ser) p=0.026. When categorized by age, both CT (Pro/Ser) and TT (Ser/Ser) alleles were significantly higher (p= 0.003 & 0.004 respectively) in cases above 40years of age. NQO1 protein was 42% reduced in buccal tissues of heterozygous (Pro/Ser) carriers, whereas a 70% reduction was observed in TT (Ser/Ser) OSF cases.
Conclusion: Our study suggests that the NQO1 C609T polymorphism confers increased risk for OSF in habitual chewers.
Introduction
Upsurge in the popularity of commercially prepared areca nut preparations (popularly known as ‘pan-masala & gutka’), as a chewing habit has contributed to a high prevalence of oral carcinogenesis in India [1-3]. Chewing of areca nut with betel quid increases the concentrations of carcinogenic nitrosamines and reactive oxygen species in the mouth [4-5]. As an early sign of damage to the oral mucosa, chewers often develop precancerous oral lesions such as leukoplakia and submucous fibrosis. Oral submucous fibrosis (OSF) is an insidious, chronic, progressive precancerous condition of the oral cavity and oropharynx with a high degree of malignant potentiality. A significant number of this precancerous condition converts into Oral Squamous Cell Carcinoma (OSCC), the rate being about 7.4-13% [6]. This disease is now a public health concern in many parts of the world including many southeast Asian countries, the United Kingdom, and South Africa though it is mainly prevalent in the Indian subcontinent among all age groups and across all socioeconomic strata [7-8]. The human NAD (P) H: quinone oxidoreductase 1 gene (NQO1; DT-diaphorase, Enzyme Commission (EC) number 1.6.99.2) occupies 17 kilobase pairs (kb) within a gene-rich region on chromosome 16 at 16q22.1 [9]. This cytosolic flavoenzyme detoxifies quinones (a large class of aromatic compounds found commonly in plants, benzene metabolites, and chemotherapies) to hydroquinones or catechols. The enzyme NAD(P)H: quinone oxidoreductase 1 (NQO1) acts as an antioxidant by catalyzing a 2-electron reduction that bypasses the need for two 1-electron reductions that can result in the production of DNA and protein-damaging reactive oxygen species. In certain conditions (e.g., the presence of myeloperoxidase or antioxidants), NQO1 can contribute to the formation of reactive oxidation species via oxidative cycling and therefore can act as a prooxidant [10]. NQO1 is constitutively expressed in most tissues including the bone marrow. NQO1 expression is thought to be highly inducible by xenobiotics with quinone moieties and is upregulated during times of oxidative or electrophilic stress.
The polymorphisms of NQO1gene have been characterized and known for about two decades [11]. A C>T substitution at position 609 of NQO1 cDNA, which codes for a Proline > Serine (Pro>Ser) change at residue 187 is associated with the risk of several cancers to date [12]. Functionally, rapid degradation of NQO1 was observed in colon cancer cells transfected with a mutant 609T vector in vitro. The presence of a Pro>Ser mutation rapidly targets the NQO1 protein for ubiquitin-mediated proteasomal degradation pathway leading to reduced NQO1 activity in these cells [13]. The prevalence of the minor T allele varies among ethnic groups (4–20%), with the highest prevalence occurring in Asian populations [14-15]. In one of our previous observational studies, we found a very high prevalence of NQO1 Pro>Ser polymorphism among both urban and rural populations of West Bengal, India, who were habitually exposed to processed areca nut and smokeless tobacco [16]. This study explores the effect of NQO1 Pro>Ser polymorphism in increasing susceptibility to Oral Submucous fibrosis among habitual areca nut and smokeless tobacco chewers.
Materials and Methods
Patients and controls
A total of 179 newly diagnosed cases of oral submucous fibrosis and 152 healthy control subjects having an oral habit for a similar period were recruited from the outpatient’s department at Dr. R Ahmed Dental College & Hospital, Kolkata, India over 3 years (2010-2013). All individuals enrolled in the study provided informed consent for their participation in the study. All participants completed a questionnaire providing information about their age, gender, ethnic origin, and use of processed areca nut products, smoking or alcohol consumption status. Written consent was obtained from all the patients after educating them about the importance and outcome of the study and all the study protocols were reviewed by an institutional ethical committee. Blood was collected from the antecubital vein from all the study participants. A biopsy sample was collected from the affected tissue. A part of the tissue was used for routine histopathological and immunohistochemical examination, while another part was used for further analysis of genetic or protein expression. NQO1 mRNA and protein expression were evaluated in 10 cases each of wild type, heterozygous and homozygous carriers. 5 normal samples with wild-type genotype were obtained from healthy volunteers during 3rd molar extraction.
Genotyping
Genotyping was carried out by polymerase chain reaction-restriction fragment length polymorphism analysis on DNA isolated from whole blood samples. The primers used were forward: 5`AAGCCCAGACCAACTTCT-3` and reverse:5`-ATTTGAATTCGGGCGTCTGCTG-3`. PCR amplification was performed by initial denaturation of 95oC for 2 mins followed by 35 cycles of 94oC for 30 secs, 60oC for 30secs, 72oC for 1 min and a final elongation of 72oC for 10 mins. Subsequently, the PCR products were digested with 20 U of HinfI (New England Biolabs, USA) for 3 hr at 37ºC and separated on a 6% polyacrylamide gel. The NQO1 wildtype allele was identified by an undigested 172 base pair (bp) PCR product. Whereas the homozygous minor and heterozygous alleles were identified by the presence of two (131 bp and 41 bp) and three (172,131 and 41 bp) bands respectively. The frequency of the NQO1 genotypes was compared in the patient and control groups.
RNA isolation from oral mucosal tissue
Total RNA was extracted using Trizol reagent (Gibco BRL, Gaithersburg, MD) according to the manufacturer’s protocol. Approximately 50 mg of tissue was collected in 1 ml Trizol reagent and homogenized using a handheld homogenizer. After the addition of 0.2 ml chloroform, the mixture was vigorously shaken for 3 min at 22°C and centrifuged at 13000 rpm for 15 min at 4°C. An equal volume of isopropanol (chilled) was added and was kept in cold condition for 10 min, RNA was precipitated by centrifugation at 13000 rpm for 10 min at 4°C. The pellet was washed twice with 70% ethanol, briefly dried under air, and dissolved in 100 µl of diethylpyrocarbonate- treated water.
Quantitative real-time PCR of NQO1 mRNA
Isolated RNA from OSF and control tissues was immediately subjected to real-time RT PCR (using Takara PrimescriptTM one-step real-time RT PCR kit) (Takara, Japan) analysis for detection of changes in expression of NQO1 mRNA according to the manufacturer’s protocol. Briefly, the PCR was carried out using the following primers Forward 5’-GGGCAAGTCCATCCCAACTG-3’ and Reverse 5′GCAAGTCAGGGAAGCCTGGA-3’ (230 bp) while for GAPDH the forward and reverse primers were 5’-ATGGGGAAGGTGAAGGTCGG-3’ and 5’-GGATGCTAAGCAGTTGGT-3’ respectively, yielding a 470 bp product. The reaction mix contained 200 ng of RNA; PrimeScript One Step Enzyme Mix; 1X One-Step Buffer; 20 µM of each of the primers and RNase Free dH2O. The real-time RT-PCR cycling program involved an initial denaturation step at 95°C for 2 min, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. Thermal cycling was performed in a BioRad iQ5 Real-Time PCR Detection System (Hercules, CA, USA).
Western blotting
The tissues were collected in PBS buffer containing protease inhibitor cocktail (Sigma-Aldrich, USA). Tissue extracts were prepared by RIPA lysis buffer [150 mM NaCl 1% Nonidet P-40 (vol/vol) 0.5% AB-deoxycholate (vol/vol) 0.1% SDS (vol/vol) 50mMTriszHCl (pH 8) 1mM DTT 1mg/ml each of leupeptin, aprotinin, and pepstatin]. The insoluble pellet was discarded, and the protein concentration was determined by using Bradford reagent (Bio-Rad). Equal amounts of protein were mixed with sample buffer (4% SDS 20% glycerol 10% 2-mercaptoethanol 0.125 M Tris-HCl), heated at 95°C for 5 min, loaded, and separated on 6% polyacrylamide- SDS gel. After separation, the proteins were blotted onto a PVDF membrane (Millipore, Billerica MA) and incubated with mouse monoclonal NQO1 (1:1000) (Cell Signalling Technology, Beverly, MA) and β-Actin Antibody (C4) (1:2000) (Santa Cruz Biotechnology, USA) as primary antibodies. Goat anti-mouse secondary antibody conjugated with ALP (1:2000 dilution) was then added and incubated for 3 hours at room temperature followed by washing in TBST six times for ten minutes each to detect the protein levels. The alkaline phosphatase-positive bands were visualized using a 5-Bromo,4-chloro,3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) (GENEI). The band’s intensities were analyzed using ImageJ (http:// rsbweb.nih.gov/ij/download.html) and expressed in densitometric units (DU).
Immunohistochemistry
Paraffin blocks each from wild type, heterozygous and homozygous mutant OSF cases were selected for subsequent immunohistochemical investigations. Briefly, dissected oral tissues from biopsy specimen were paraffin-embedded and 4-5µm thickness sections were collected on poly-L-lysine coated slides, after paraffin removal by xylene and rehydration, the slides were treated with citrate buffer for unmasking the antigen. Further immunostaining was performed using the Novolink polymer detection system (Leica Microsystems, Switzerland). The endogenous peroxidase and protein were blocked using supplied blockers. The expression of NQO1 protein was detected using NQO1 monoclonal antibody (Cell signaling technologies, Beverly, MA). After post-primary blocking the sections were incubated with Novolink polymer and were then developed with DAB using supplied substrate buffer. The sections were then counterstained with hematoxylin and were observed under LEICA DM 3000 microscope ((Leica Microsystems, Switzerland).
NQO1 activity assay from affected and adjacent normal tissue
NQO1 activity was measured from a part of biopsy tissues obtained from three OSF cases each with either heterozygous (CT) or homozygous (TT) OSF, OSF associated with malignancy (n=3) and healthy tissue (n=3) collected during 3rd molar extraction. Total protein was isolated and determined by the Biorad Protein assay kit (Hercules, CA, USA) using bovine serum albumin as a standard. The assay was performed as described by Ernster (1967) [17]. and modified by Benson et al. (1980) [18] using DCPIP as a substrate. The reaction mix contained 25 mM Tris (pH 7.4), with or without 0.07% bovine serum albumin (w/v) (Sigma-Aldrich, USA) as an activator, 200µM NADH (Sigma-Aldrich, USA) and 40µM DCPIP (Sigma-Aldrich, USA), and assays were carried out in different concentration of Dicumarol (Calbiochem, USA). NQO1 activity is described as the dicumarol inhibitable decrease in absorbance at 600 nm with DCPIP as a substrate and is expressed in nanomoles of DCPIP reduced per minute per milligram of protein.
Single-cell gel electrophoresis
For single cell gel electrophoresis or ‘COMET’ assay, individuals were asked to rinse their mouths thoroughly for 2min with tap water. The exfoliated buccal cells were collected from one or both cheeks from the controls, and in the area with lesion from the cases, depending on the region where the lesion was located, including cheek, soft and hard palate, dorsal, ventral, and lateral surfaces of the oral cavity. Typically, sites with a tough, leathery texture of mucosa, blanching of mucosa (persistent, white, a marble-like appearance which may be localized, diffuse or reticular), quid-induced lesions (fine, white, wavy, parallel lines that do not overlap or criss-cross, are not elevated and radiate from a central erythematous area) were selected. Comet assay was performed and analyzed as described previously [19].
Statistical analysis
The calculation of genotypic and allele frequencies for cases and controls and association between polymorphisms of NQO1 C609T with the risk of oral submucous fibrosis among the total and stratified population was estimated by computing odds ratio (OR) and calculating 95% Confidence Interval (CI) using a chi-square Table Analysis and Yates corrected P-value was taken for significance. The expression of NQO1 protein was analyzed densitometrically using Image J software after proper calibrations. The difference in protein expression was analyzed by the Student “t” test, p-value <0.05 was significant.
Results
The study population was stratified according to Gender (Male and Female) and Median age (40 years) as presented in Table 1.
| OSF cases (179) | Healthy Controls (152) | |
| Age | ||
| Range | 14-73 | 18-78 |
| Mean | 40 | 37 |
| Gender | ||
| Male | 118 (65.9%) | 74 (48.6%) |
| Female | 61 (34.1%) | 78 (52.4%) |
| Above 40 yrs | 118 (65.9%) | 94 (61.8%) |
| Below 40 yrs | 61 (34.1%) | 58 (39.1%) |
We found that males (65.9%) constituted a greater percentage of the study sample and that most of the OSF cases were in the age group of > 40 years (65.9%).
Distribution of Genotypes of NQO1 C609T polymorphism among the cases and controls
Wild type C/C allele, heterozygous C⁄T allele, and homozygous mutant T/T alleles were determined by PCR- RFLP method (Figure 1A).
Figure 1. Effect of Various Genotypes on Expression Patterns of NQO1 and Buccal Tissue Architecture. A) Genotype determination by restriction fragment length polymorphism following PCR amplification (PCR-RFLP). The size of digested products is shown in the figure. B) Increased mRNA expression wrt GAPDH of NQO1 in OSF cases compared to controls indicating transcriptional up-regulation in affected tissues. C) Expression levels of NQO1 protein in different genotypic conditions in OSF cases. D) Immunohistochemical localization shows the high basal level expression of NQO1 especially along the rete-ridges and submucosal cells lining epithelium in OSF cases with Pro/Pro allele (ii) compared to controls (i). Little or no detectable expression is observed in heterozygous (Pro/Ser) (iii) or homozygous (Ser/Ser) (iv) oral mucosal tissue. E) DNA damage as evident from ‘COMET’ assay performed on exfoliated buccal tissue from OSF cases with different genotypes. DNA disintegration worsens with heterozygous or homozygous mutant conditions.
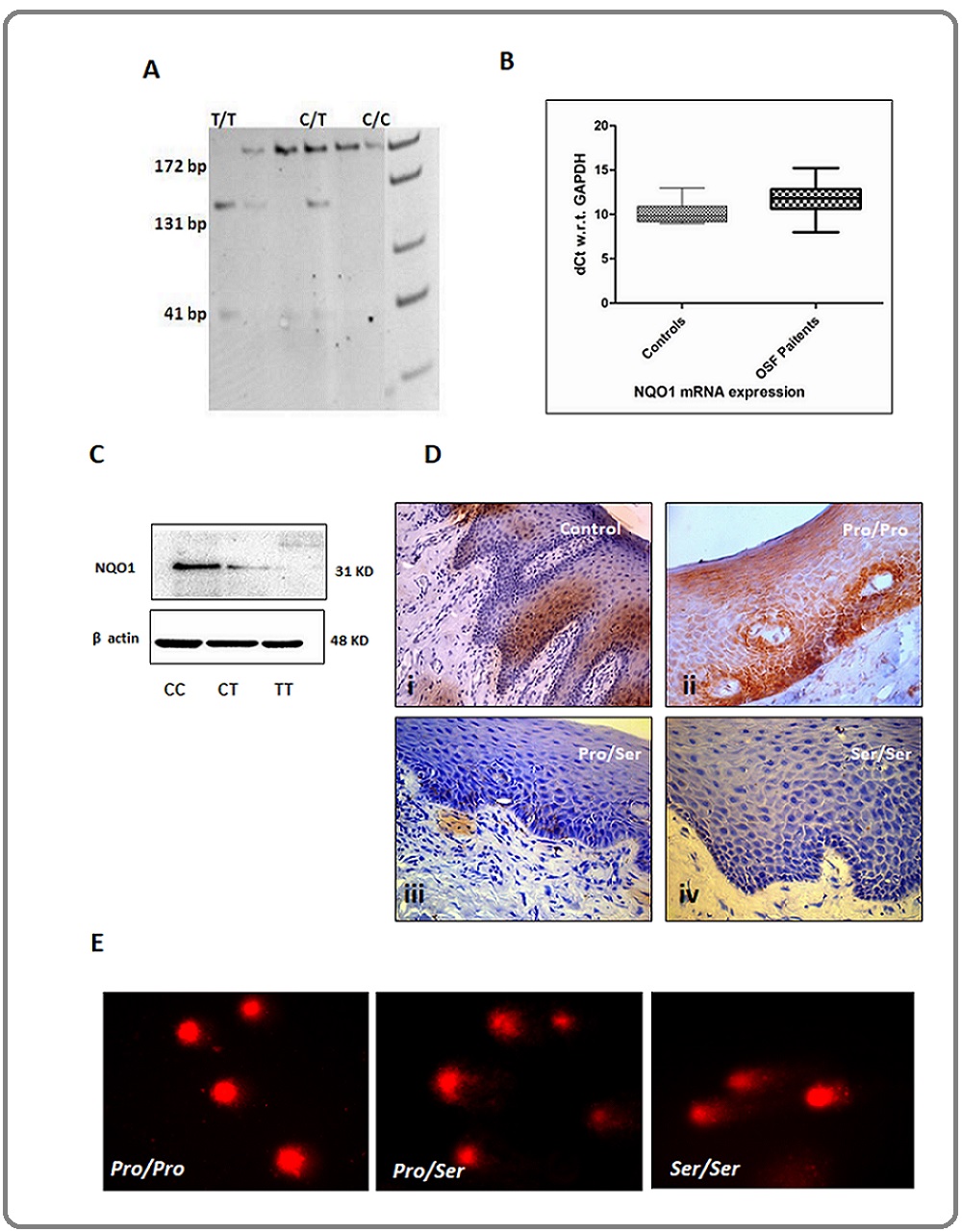
Sequencing of random samples was done to check the genotype and compared to those obtained by PCR-RFLP to rule out experimental errors. The distribution of genotypes among the study samples (cases and control) is presented in Table 2.
| Cases | Normal | OR* | 95% CI** | P-Value | |
| Total population | n=179 | n=152 | |||
| Pro/Pro | 80 (44.7) | 77 (50.65) | ref | ||
| Pro/Ser | 67 (37.43) | 62 (40.78) | 1.056 | 0.664-1.679 | 0.913 |
| Ser/Ser | 32 (17.8) | 13 (8.5) | 2.369* | 1.167-4.804 | 0.026 |
| Below 40 yrs | n=128 | n=96 | |||
| Pro/Pro | 60 (46.87) | 33 (34.04) | ref | ||
| Pro/Ser | 48 (37.5) | 54 (56.3) | 0.493 | 0.27-0.87 | 0.024 |
| Ser/Ser | 20 (15.6) | 9 (9.5) | 1.185 | 0.49-2.85 | 0.893 |
| Above 40 yrs | n=51 | n=56 | |||
| Pro/Pro | 20 (39.2) | 43 (76.7) | ref | ||
| Pro/Ser | 19 (37.2) | 9 (16.07) | 4.5* | 1.77-11.61 | 0.003 |
| Ser/Ser | 12 (23.5) | 4 (7.1) | 6.4* | 1.92-21.35 | 0.004 |
| Male | n=118 | n=74 | |||
| Pro/Pro | 52 (44.06) | 30 (40.5) | ref | ||
| Pro/Ser | 46 (38.9) | 38 (51) | 0.698 | 0.376-1.297 | 0.329 |
| Ser/Ser | 20 (16.9) | 6 (8.1) | 1.923 | 0.711-5.59 | 0.3 |
| Female | n=61 | n=78 | |||
| Pro/Pro | 28 (45.9) | 32 (41.02) | ref | ||
| Pro/Ser | 21 (34.4) | 39 (50) | 0.615 | 0.297-1.276 | 0.265 |
| Ser/Ser | 12 (11.4) | 7 (8.9) | 1.95 | 0.95-5.15 | 0.322 |
* Odds Ratio; ** Confidence interval
Overall, the frequency of homozygous mutant type (18%) was significantly higher in OSF cases than controls (8.5%) [OR of 2.369 (1.167-4.804) P=0.026]. When stratified according to age, both the heterozygous carrier and homozygous mutant variant were found to be higher in cases above 40 years of age compared to controls OR 4.5 (1.77-11.61) P=0.003 and OR 6.4 (1.92-21.35) P=0.004
respectively.
NQO1 expression and epithelial DNA disintegration in different genotypes
NQO1 mRNAexpression was higher in cases with either genotype compared to controls indicating transcriptional up-regulation of NQO1 in tissues (Figure 1B). In general, a significantly decreased mean expression for NQO1 protein was observed in buccal mucosal tissues of cases carrying heterozygous (42%) or homozygous (70%) genotypes (p=0.0055 & p=0.0001 respectively) indicating NQO1 degradation in homozygous/heterozygous mutant cases is a post-transcriptional event (Table 3) (Figure 1C).
| Genotype | P value | ||
| Pro/Pro Vs Pro/Ser | Pro/Pro | Pro/Ser | 0.0055 |
| 1.4875±0.8046 | 0.8789±0.5969 | ||
| Pro/Pro Vs Ser/Ser | Pro/Pro | Ser/Ser | 0.0001 |
| 1.4875±0.8046 | 0.4297±0.2492 |
Histochemical localization of NQO1 protein in oral epithelial cells (Figure 1D) is concurrent with western-blots and shows high concentrations of NQO1 expression in epithelium of the tissue sections obtained from OSF cases with wild type Pro/Pro genotype compared to almost focal expression in the normal healthy epithelium. Sections show increased NQO1 expression around the basal cell layer in wild-type cases. NQO1 expression in heterozygous Pro/Ser OSF cases showed faint localizations whereas cases with homozygous mutant trait showed almost no NQO1 expression in the basal cells or elsewhere.
Comparative analysis of single-cell gel electrophoresis of exfoliated buccal cell collected from the site of the lesion, reveals a higher degree of disintegrated DNA in heterozygous and homozygous mutant OSF cases (Figure 1E) compared to wild types.
Lower NQO1 activity in OSF and OSF associated with malignancy tissues compared to their adjacent tissue:
Figure 2A represents the NQO1 activity (%) in normal, OSF, and OSF with malignancy (OSF+MAL) tissues in the presence or absence of BSA. When compared to the OSF tissue alone there was an about 2-fold decrease in activity (p=0.028) with or without BSA.
Figure 2. A) Graphical Presentation of NQO1 Activity in Presence of Dicoumarol in Normal, OSF, and OSF with Malignancy (OSF+MAL) Tissue Extract in the Absence (gray bars) or Presence (black bars) of BSA as an Activator. B) NQO1 activity as measured by reduction of DCPIP in presence of dicoumarol with or without activator. The OSF with malignant lesion cases (OSF+MAL) carrying Ser/Ser allele presents reduced or no activity of NQO1 compared to normal or OSF cases with no malignancy. MAL - Malignancy.
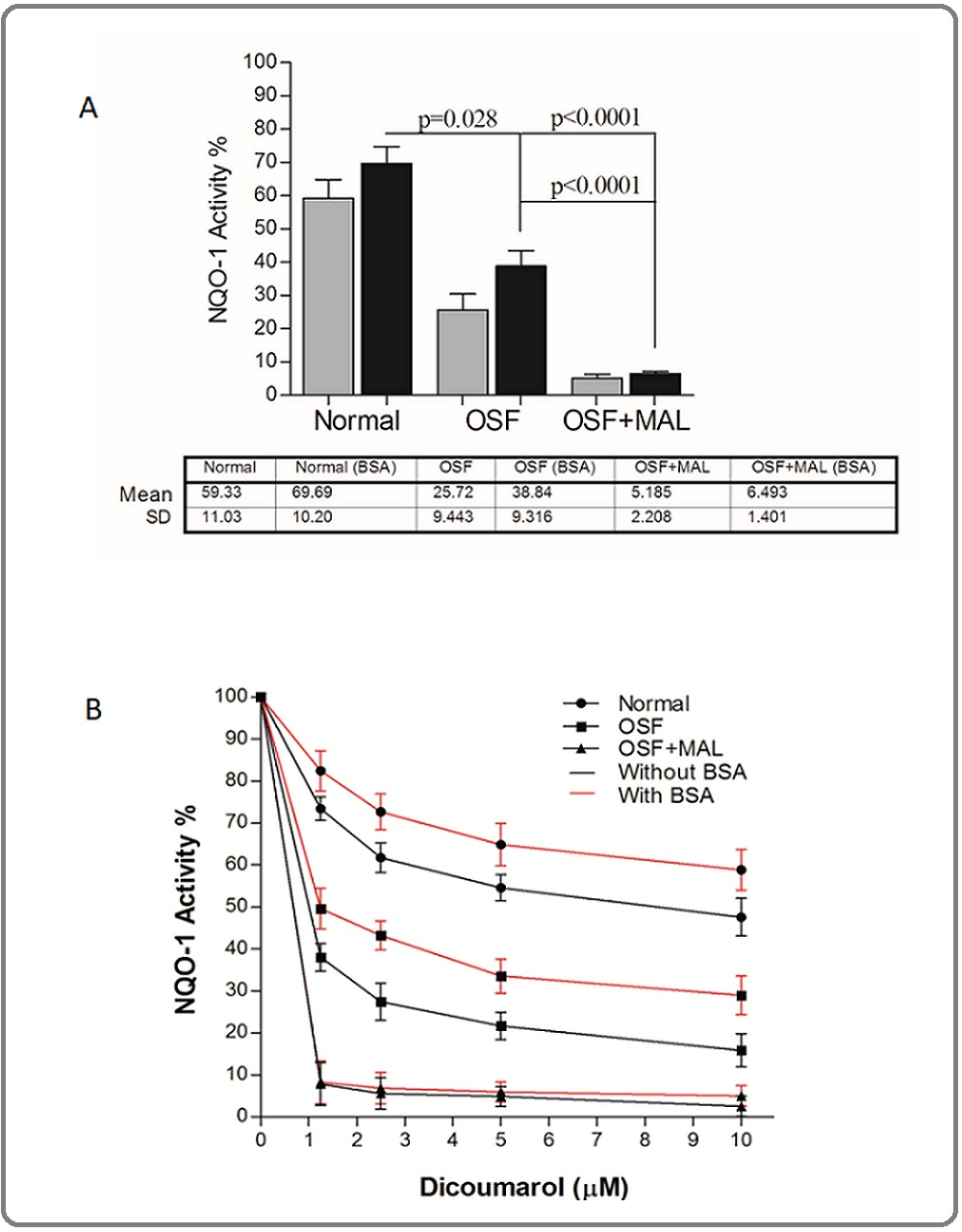
This was about a 10-fold decreased in OSF associated with malignant tissues (p< 0.0001) thus explaining insignificant NQO1 activity. Figure 2A presents kinetics of NQO1-mediated reduction of DCPIP as a measure of NQO1 activity. The Vmax and Km of NQO1 at different concentrations of dicoumarol inhibitor in Normal (n=10), OSF (n=10 from each genotype), and OSF associated with malignancy cases with homozygous mutant genotype (n=4) are presented in supplementary table 1. With an increase in the concentration of Dicoumarol (0, 1.25, 2.5, 5, and 10 µM) there was a uniform decrease in Vmax of NQO1 but no significant change in Km value was detected either in the presence or absence of the activator (BSA). This suggests there is a non-competitive inhibition of the NQO1 activity in the presence of the inhibitor.
Discussion
Oral submucous fibrosis is a potentially malignant condition affecting any part of the oral cavity and pharynx, which could subsequently develop into oral cancer [20]. The carcinogenic turnover of OSF may serve as a very important model to study various molecular changes that are taking place during oral carcinogenesis. In the present study, we have tried to understand the pathogenesis and carcinogenesis of OSF in the light of NQO1 C609T polymorphism. In OSF mucosal tissues from cases carrying Ser/Ser genotype, either traces or no NQO1 protein was detected, this might be due to ubiquitin-mediated proteasomal degradation as discussed earlier [13]. In the present study, we observed a significantly higher number of subjects carrying the NQO1 homozygous mutant allele genotype in cases suffering from oral submucous fibrosis as compared to the control group. Mostly male cases with OSF reported to our outpatient department (OPD) compared to the female cases with a median of 40 years of age. Cases above 40 yrs showed a greater association with the risk of the disease. This result is in line with previous investigations that showed an association between the NQO1 homozygous mutant genotype and other tumor types such as urothelial cancer, renal cancer, leukemia, lung cancer, and cutaneous basal cell carcinoma [21-25]. Overall, the epithelial NQO1 expression in OSF cases with wildtype NQO1 genotype was higher compared to the normal epithelium. Since, NQO1 expression is ‘switched on’ due to oxidative stress, higher exposure to oral chewing habit may lead to higher expression as a result of defensive mechanisms. Besides, we also observed a reduction of epithelial NQO1 expression in either heterozygous (CT) (42%) or in homozygous (TT) (70%) OSF cases. The NQO1 activity either in OSF buccal tissues with CT/TT genotypes or OSF associated with malignancy showed similarly reduced activity compared to the wild-type control buccal tissue. The ubiquitin-mediated proteasomal degradation pathway leading to reduced NQO1 availability or activity due to the presence of a Pro>Ser mutation might be responsible for such observation and further studies with a larger sample size need to be done. The mechanism underlying the correlation of NQO1 C609T polymorphism with the increased risk for developing various tumors likely resides in the different enzyme activities encoded by the NQO1 alleles. There were two major limitations in our study, firstly, the availability of biopsy tissues especially from OSF cases or OSF associated with malignancy cases for any functional studies, that could have been more suggestive of the mechanism of carcinogenesis. Secondly, lack of follow-up studies due to the unavailability of study patients.
Several cancer chemotherapeutic drugs have been developed targeting NQO1 [26]. Since NQO1 C609T polymorphism is found to be positively associated with many solid as well as blood malignancies, therefore, a practical approach for cost-effective tumor screening may be designed taking other such gene polymorphisms, especially the metabolic genes [27] into account before starting a treatment regimen.
Acknowledgments
The authors mourns the death of Dr. Keya Chaudhuri and sincerely acknowledge her contributions as the principal investigator of this study. The study was supported by the Council of Scientific and Industrial Research, Govt. of India. SM is grateful to CSIR for the Senior Research Fellowship. AK is grateful to ICMR for the Senior Research Fellowship.
References
- Oral submucous fibrosis: etiology, pathogenesis, and future research Rajendran R. Bull World Health Organ.1994;72(6):985-996.
- Oral submucous fibrosis: A global challenge. Rising incidence, risk factors, management, and research priorities Ray Jay Gopal, Chatterjee Raghunath, Chaudhuri Keya. Periodontology 2000.2019;80(1). CrossRef
- Medicinal management of oral submucous fibrosis in the past decade- A systematic review More Chandramani B., Jatti Patil Deepa, Rao Naman R.. Journal of Oral Biology and Craniofacial Research.2020;10(4). CrossRef
- Role of lime in the generation of reactive oxygen species from betel-quid ingredients. Nair U J, Obe G, Friesen M, Goldberg M T, Bartsch H. Environmental Health Perspectives.1992;98. CrossRef
- Evaluation of cytotoxicity of areca nut and its commercial products on normal human gingival fibroblast and oral squamous cell carcinoma cell lines Adil Nurmeen, Ali Hamad, Siddiqui Amna Jabbar, Ali Arslan, Ahmed Ayaz, El-Seedi Hesham R., Musharraf Syed Ghulam. Journal of Hazardous Materials.2021;403. CrossRef
- Oral submucous fibrosis: an unusual disease Aziz SR. Journal of the New Jersey Dental Association.1997;68(2):17-19.
- Global epidemiology of areca nut usage Gupta P. C., Warnakulasuriya S.. Addiction Biology.2002;7(1). CrossRef
- Areca nut use: an independent risk factor for oral cancer Warnakulasuriya S.. BMJ.2002;324(7341). CrossRef
- Human dioxin-inducible cytosolic NAD(P)H:menadione oxidoreductase. cDNA sequence and localization of gene to chromosome 16 Jaiswal AK, McBride OW, Adesnik M, Nebert DW. The Journal of biological chemistry.1988;263(27):13572-13578.
- NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms Ross David, Kepa Jadwiga K, Winski Shannon L, Beall Howard D, Anwar Adil, Siegel David. Chemico-Biological Interactions.2000;129(1-2). CrossRef
- Characterization of a polymorphism in NAD(P)H: quinone oxidoreductase (DT-diaphorase) Traver RD, Siegel D, Beall HD, Phillips RM, Gibson NW, Franklin WA, Ross D. British Journal of Cancer.1997;75(1). CrossRef
- The NQO1 polymorphism C609T (Pro187Ser) and cancer susceptibility: a comprehensive meta-analysis Lajin B, Alachkar A. British Journal of Cancer.2013;109(5). CrossRef
- Rapid Polyubiquitination and Proteasomal Degradation of a Mutant Form of NAD(P)H:Quinone Oxidoreductase 1 Siegel David, Anwar Adil, Winski Shannon L., Kepa Jadwiga K., Zolman Kathryn L., Ross David. Molecular Pharmacology.2001;59(2). CrossRef
- Ethnic variation in the prevalence of a common NAD(P)H quinone oxidoreductase polymorphism and its implications for anti-cancer chemotherapy Kelsey KT, Ross D, Traver RD, Christiani DC, Zuo Z-F, Spitz MR, Wang M, Xu X, Lee B-K, Schwartz BS, Wiencke JK. British Journal of Cancer.1997;76(7). CrossRef
- NAD(P)H:quinone oxidoreductase: polymorphisms and allele frequencies in Caucasian, Chinese and Canadian Native Indian and Inuit populations Gaedigk Andrea, Tyndale Rachel F., Jurima-Romet Malle, Sellers Edward M., Grant Denis M., Leeder J Steven. Pharmacogenetics.1998;8(4). CrossRef
- Comparative distribution of Lysyl Oxidase (G473A) and NQO1 (C609T) polymorphism among tea-garden workers (habitual chewers of betel quid) of Darjeeling district and Kolkata city of West Bengal Chaudhuri Keya, Ray JayGopal, Mukherjee Sanjit, Mahato Basudev, Sripathi Rao BH. Contemporary Clinical Dentistry.2013;4(4). CrossRef
- [56] DT diaphorase. Methods in Enzymology Ernster L, Ronald W, Estabrook MEP. Academic Press; 1967;10:309-317.
- Increase of NAD(P)H:quinone reductase by dietary antioxidants: possible role in protection against carcinogenesis and toxicity. Benson A. M., Hunkeler M. J., Talalay P.. Proceedings of the National Academy of Sciences.1980;77(9). CrossRef
- Comparative evaluation of genotoxicity by micronucleus assay in the buccal mucosa over comet assay in peripheral blood in oral precancer and cancer patients Katarkar Atul, Mukherjee Sanjit, Khan Masood H., Ray Jay G., Chaudhuri Keya. Mutagenesis.2014;29(5). CrossRef
- Malignant Conversion of Oral Submucous Fibrosis in Surgically Treated Case Arora Rajendra. JOURNAL OF CLINICAL AND DIAGNOSTIC RESEARCH.2014. CrossRef
- Human NAD(P)H:quinone oxidoreductase 1 (NQO1) and sulfotransferase 1A1 (SULT1A1) polymorphisms and urothelial cancer risk in Taiwan Wang Yuan-Hung, Lee Ying-Huei, Tseng Po-Tsang, Shen Cheng-Huang, Chiou Hung-Yi. Journal of Cancer Research and Clinical Oncology.2007;134(2). CrossRef
- Increased frequency of a null-allele for NAD(P)H: quinone oxidoreductase in patients with urological malignancies Schulz Wolfgang A., Krummeck Angelika, Rosinger Ingrid, Eickelmann Peter, Neuhaus Christine, Ebert Thomas, Schmitz-Dräger Bernd J., Sies Helmut. Pharmacogenetics.1997;7(3). CrossRef
- NQO1 Polymorphisms and De Novo Childhood Leukemia: A HuGE Review and Meta-Analysis Guha N., Chang J. S., Chokkalingam A. P., Wiemels J. L., Smith M. T., Buffler P. A.. American Journal of Epidemiology.2008;168(11). CrossRef
- The NQO1 C609T polymorphism and risk of lung cancer: a meta-analysis Liu Y, Zhang D. Asian Pac J Cancer Prev.;12(11):3091-3095.
- Association of NAD(P)H:quinone oxidoreductase (NQO1) null with numbers of basal cell carcinomas: use of a multivariate model to rank the relative importance of this polymorphism and those at other relevant loci Clairmont A.. Carcinogenesis.1999;20(7). CrossRef
- Implications of NQO1 in cancer therapy Oh Eun-Taex, Park Heon Joo. BMB Reports.2015;48(11). CrossRef
- CYP1AI and CYP2E1 gene polymorphisms may increase susceptibility to Oral Submucous Fibrosis among betel quid chewers of Eastern India Chaudhuri Susri Ray, Mukherjee Sanjit, Paul Ranjan Rashmi, Haldar A., Chaudhuri Keya. Gene.2013;513(2). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2021
Author Details